Figure 5.
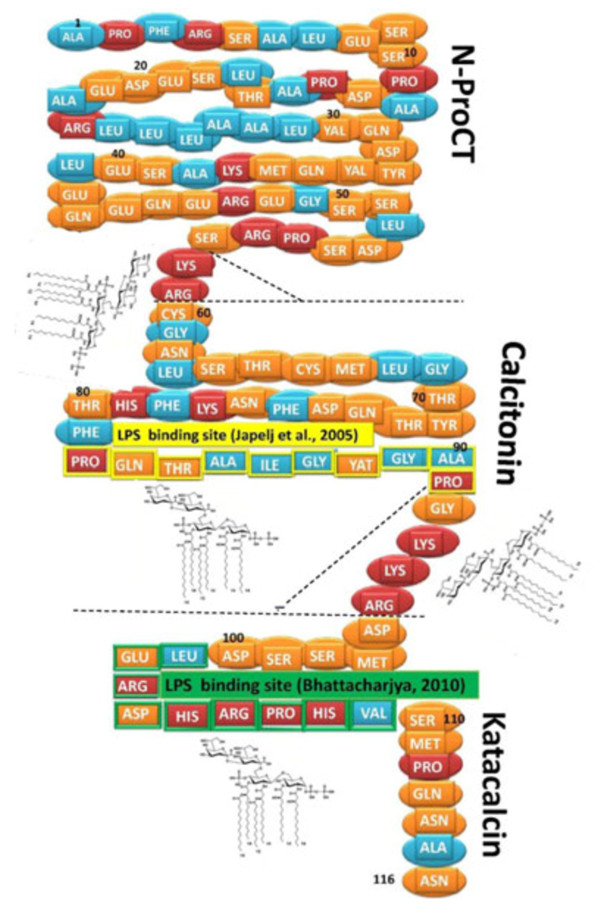
Putative LPS binding sites on PCT molecule. Proposed LPS binding sites include: i) 2–3 cationic aminoacids within a cluster of four (aminoacids 58–59 and aminoacids 93–95), ii) a cluster of hydrophobic residues encompassed by basic aminoacids (82–92), iii) a group of positively charged central residues with hydrophobic aminoacids in the periphery (101–109). Hydrophobic aminoacids in blue, cationic aminoacids in red and other aminoacids in orange. The LPS binding sites suggested by Japelj [14] and Bhattacharjya [15] are indicated. Close to the proposed LPS binding sites, a deep rough LPS chemical structure is showed. Flat dashed lines indicate the limits of the three post-translational processing products (N-ProCT, calcitonin and katacalcin) of procalcitonin, while dashed forks encompass the peptides cleaved during post-translational processing [1,3].
