Abstract
Background
Cleft lip only (CLO) and cleft lip and palate (CLP) are commonly regarded as variants of the same defect and traditionally combined to form the single group of cleft lip with or without cleft palate (CL/P) prior to analysis. However, recent data have suggested that at least a subgroup of isolated CLO may be etiologically distinct from isolated CLP.
Methods
To explore fetal genetic risk of isolated CLO separately from isolated CLP, we performed a sub-phenotype analysis using two population-based studies of clefts in Scandinavia. One hundred and twenty-one isolated CLO, 190 isolated CLP, and 592 control triads were available from Norway(1996–2001), and a further 76 isolated CLO and 107 isolated CLP triads were available from Denmark (1991–2001). Genotypes for 1315 SNPs in 334 autosomal cleft candidate genes were analyzed using two complementary statistical methods, TRIMM and HAPLIN, to look for genetic associations across the two national samples.
Results
Both TRIMM and HAPLIN identified strong associations between FGF12 and isolated CLO in both populations. In addition, only TRIMM identified associations with IRF6 and VCL, and only HAPLIN found an association with CX43. When analyses were repeated on the larger sample of isolated CLP, no significant associations were found with FGF12, IRF6, VCL or CX43.
Conclusions
Despite some inconsistency in the pattern of associations across the two populations, the associations themselves were phenotype-specific. While both IRF6 and FGF12 have previously shown strong associations with isolated CL/P, the associations with VCL and CX43 are novel and warrant further investigation in other isolated CLO samples.
Keywords: Birth defects, Orofacial cleft, Cleft lip, Cleft palate, Genetic epidemiology
INTRODUCTION
Cleft lip defects comprising CLO and CLP are usually considered variants of the same defect, with bilateral CLP representing the most severe form. They are therefore routinely lumped together to form the single group of CL/P prior to analysis (Mitchell et al., 2002). Cleft palate (CP), on the other hand, is well established as a separate entity from cleft lip defects based on embryological and epidemiological evidence (Marazita, 2002; Mossey and Little, 2002).
One of the ways to determine whether CLO and CLP have different underlying genetic predispositions is to compare the strength of the recurrence relative risk of CLO with that of CLP in first-degree relatives. The recurrence relative risk for CLO is the risk of CLO in those with a sibling (or parent) with the defect divided by the risk of CLO in those without a sibling (or parent) with the defect. The cross-over recurrence relative risk is the risk of CLO given a family member with CLP divided by the risk of CLO without a family member with CLP. If the data suggest a higher recurrence relative risk of the same defect compared with the risk of the alternate defect (the cross-over risk), then this would suggest non-identical genetic etiologies for CLP and CLO. In a Norwegian study (Sivertsen et al., 2008), no statistical difference in these two relative risks was observed, which may be due to the small number of recurrent cases available for comparisons and a lack of statistical power to identify a true difference. By contrast, a Danish cohort study approximately three times the size of the Norwegian studyuncovered important differences between CLO and CLP (Grosen et al., 2009). The sibling-sibling recurrence risk of isolated CLO (given isolated CLO) was 1.4 (95% confidence interval: 1.0–1.9) compared with the cross-over recurrence risk of CLP of 1.0 (95% CI: 0.6–1.4). The corresponding recurrence risk of isolated CLP (given isolated CLP) was 2.9 (95% CI: 2.4–3.6) compared with 0.7 (95% CI: 0.5–1.1) for the crossover recurrence risk of isolated CLO.
Another study to systematically examine CLO as a separate entity from CLP was a population-based assessment of the distribution of these two cleft types by parental age, baby’s sex, hospital characteristics, region, and mother’s marital status (Harville et al., 2005). When hospital and national birth registry data were combined, 17% of CLP infants were found to have at least one other non-cleft defect compared with only9% of CLO infants. This difference is consistent with the notion that CLP represents a more severe form of CLO. However, the data also highlighted several qualitative differences between CLO and CLP which are not easily explained bydisease severity alone. There was, for instance, a stronger male predominance among CLP infants compared with CLO infants, and the risk of CLO but not CLP was increased for twins and infants whose parents were first cousins. Furthermore, in a large study of anomalies associated with clefting (Genisca et al., 2009), other major malformations were found far more commonly in CLP cases than in CLO cases, suggesting either a greater load of genetic factors, some of which might also influence other developmental processes, or that at least subsets of CLP and CLO cases have separable etiologies.
Molecular data also support etiologic differences between a subset of CLO and CLP. We have recentlyidentified a common SNP (rs642961) within a highly conserved enhancer element for the interferon regulatory factor 6 (IRF6) gene that is strongly associated with isolated CL/P (Rahimov et al., 2008). In that study, there was a clear separation of risk and transmission pattern with this SNP for isolated CLO compared with isolated CLP. Results for rs642961 were most significant for families in which affected individuals had isolated CLO, although at least one additional still unidentified variant is likely present in at least Asian populations (Rahimov et al., 2008), as would be supported by the recent modeling studies of Dickson and colleagues (Dickson et al., 2010). Finally, a similar sub-phenotypic specificity was apparent in our recent genome-wide linkage scan (Marazita et al., 2009), where results for the IRF6 region were most significant for isolated CLO.
Although the strong association of IRF6 with isolated CL/P is now well established (Jugessur et al., 2008; Rahimov et al., 2008), only a handful of studies have examined the effects of IRF6 variants on the risk of isolated CLO. In addition to the widespread practice of combining CLO with CLP prior to analysis, this may be due to the disincentive caused by the loss of statistical power in splitting CL/P into two smaller subgroups and the ensuing need to correct for multiple testing in yet another cleft subgroup. However, to confirm that observed differences between CLO and CLP are the result of distinct genetic predispositions rather than chance, more studies are needed that target isolated CLO specifically.
We screened for genetic associations among a custom panel of 1315 SNPs in 334 autosomal candidate genes for clefting using case-parent triads of isolated CLO from two nationwide case-control studies of orofacial clefts in Scandinavia (Norway and Denmark). To verify that any detected association is specific for isolated CLO, we repeated the analyses in the larger sample of isolated CLP triads from each of these two populations.
METHODS
Study populations
One hundred and twenty-one isolated CLO, 190 isolated CLP, and 592 control offspring-parent triads were available for analysis from a population-based case-control study of orofacial clefts carried out in Norway (1996–2001). In addition, 76 isolated CLO and 107 isolated CLP triads were available from a population-based study of orofacial clefts in Denmark (1991–2001). The patients from Norway were examined by a plastic surgical team using a classification system based on a modification of the “striped-Y of Kernahan” (Kernahan, 1971. Plast. Reconstr. Surg. 47(5): 469) and the RPL numerical coding by Schwartz (Schwartz et al. 1993. Cleft Palate Craniofac. J. 30(3):330). Details on study design and participants have been given elsewhere (Bille et al., 2005; Wilcox et al., 2007). Of the 121 Norwegian isolated CLO triads, 13 had at least one missing family member, bringing the number of complete triads to 108. Of the 190 Norwegian isolated CLP triads, 140 were complete. For the same reason, only 55 of the original 76 Danish isolated CLO triads and 95 of the original 107 Danish isolated CLP triads were complete.
Data analysis
The specific protocol used for the selection of candidate genes and SNPs has been detailed in (Jugessur et al., 2009b). Briefly, a custom panel of 1536 SNPs in 357 candidate genes for orofacial clefts was designed by Illumina (http://www.illumina.com; San Diego, CA) and genotyped by the Center for Inherited Disease Research (CIDR; http://www.cidr.jhmi.edu) at the Johns Hopkins University (Baltimore, Maryland). After data-cleaning, genotypes for 1315 SNPs in 334 autosomal genes were available for the current analysis. We used two complementary statistical methods, Triad Multi-Marker (TRIMM) (Shi et al., 2007) and HAPLIN (Gjessing and Lie, 2006), to screen for genetic associations across the Norwegian and Danish isolated CLO and isolated CLP samples, respectively. Both methods were designed to detect multi-marker transmission distortion in offspring-parent triads, but they accomplish this in different ways. For instance, TRIMM is non-parametric and resists bias due to population structure. It can handle deviation from Hardy-Weinberg equilibrium (HWE), multiple SNPs, missing SNPs, and non-negligible recombination rates. Since TRIMM does not attempt to infer haplotypes, it is computationally efficient. When applied to a set of SNPs within a gene, TRIMM accounts for within-gene SNP correlations by permuting alleles at all SNPs simultaneously(i.e., by permuting the case and complement labels). HAPLIN, in contrast, is parametric and estimates the full haplotype distribution over a set of SNPs and also estimates relative risks associated with each haplotype. By using a rich maximum likelihood model, it produces a complete description of the “risk structure” over the set of haplotypes in a region. HAPLIN requires HWE, assumes no recombination, and is computationally more demanding. In the current analyses, we used only three SNPs in HAPLIN for a sliding-window haplotype analysis because of the small size of the isolated CLO and isolated CLP samples. Longer window-lengths require accounting for many rare haplotypes.
Although the Norwegian and Danish populations are both geographically and ancestrally closely related to one another, as recently confirmed through principal component analysis of 3000 European individuals (Novembre et al., 2008), we nevertheless chose to perform TRIMM and HAPLIN analyses separately on the Norwegian and Danish isolated CLO samples, because our previous analysis of fetal gene effects in the same two populations revealed evidence of across-population differences (Jugessur et al., 2009b). P-values from these analyses were displayed using a Schweder-Spjøtvoll plot (Schweder and Spjøtvoll, 1982), which is a simple graphical procedure for the simultaneous evaluation of many tests. If none of the genes is truly associated with disease, p-values would fall along a sloping line that represents the expected uniform distribution under the null (of no association); they would otherwise tend to fall below the sloping line for genes that are truly associated with disease.
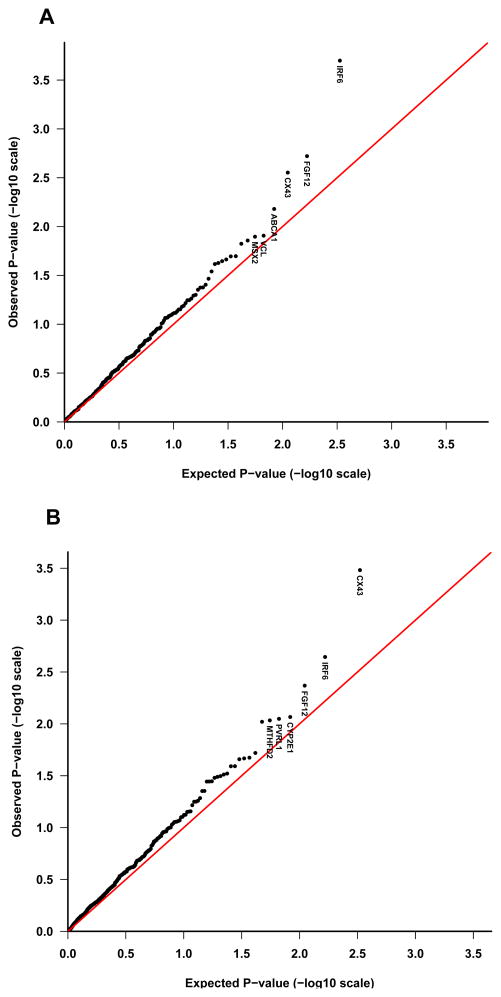
To visually inspect whether the TRIMM and HAPLIN analyses generated more significant results than expected by chance, we generated quantile-quantile (QQ) plots using p-values from the Norwegian and Danish analyses after they were combined using Fisher’s method (Fisher, 1958). If the distribution of p-values follows the null (uniform) distribution, points in the QQ plot would be close to the diagonal line. Conversely, large-effect susceptibility loci would fall above the line representing the uniform distribution, at the highly significant end of the distribution.
To estimate the relative risk associated with a candidate risk haplotype, we used TRIMM to identify risk haplotype-tagging alleles with a p-value cutoff of 0.1. That is, we nominate the risk haplotype-tagging alleles using SNPs with a p-value less than 0.1 based on individual Z-scores. We then used TRIMMest (Shi et al., 2009b), a statistical method tailored for family-based association analysis, to estimate the relative risk of the identified risk haplotypes. TRIMMest fits log-linear models to estimate risk associated with a candidate risk haplotype relative to the aggregate of other haplotypes. We fit a log-additive risk model without any assumption on the parental mating type distribution, a procedure that resists bias due to population stratification.
Finally, to verify that the associations detected are specific for isolated CLO, TRIMM and HAPLIN analyses were repeated on the larger samples of isolated CLP from the two populations.
Software for implementing TRIMM, HAPLIN and TRIMMest are available for the R computing environment (R Development Core Team, 2006) from our web sites (TRIMM and TRIMMest: http://www.niehs.nih.gov/research/atniehs/labs/bb/staff/weinberg/index.cfm#downloads; HAPLIN: http://www.uib.no/smis/gjessing/genetics/software/haplin).
Study approval
Approval for this study was obtained from the Norwegian Data Inspectorate, the Regional Committee on Research Ethics for Western Norway, and the respective Institutional Review Boards of the US National Institute of Environmental Health Sciences (NIH/NIEHS) and the University of Iowa. For the Danish orofacial clefts study, approval was obtained from the Danish National Committee on Biomedical Research Ethics. Clinicopathological information from all participating families and biologic specimens for DNA extraction were obtained with the written informed consent of the mothers and fathers. All aspects of this research were in compliance with the tenets of the Declaration of Helsinki for human research (http://www.wma.net).
RESULTS
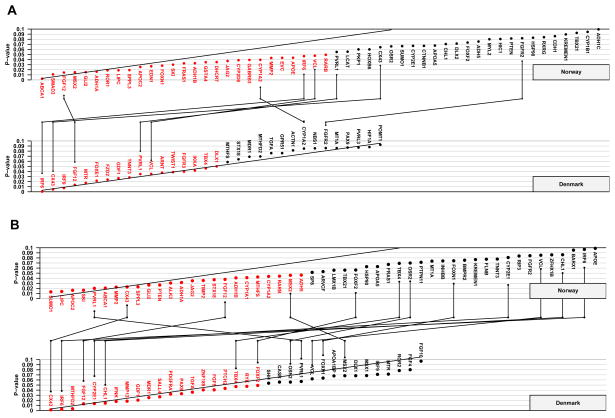
The sample distribution according to cleft type, gender, and population is provided in Table 1. Figure 1 shows Schweder-Spjøtvoll plots for all genes that had p-values ≤ 0.1 from the TRIMM and HAPLIN analyses of the Norwegian and Danish isolated CLO samples, respectively. Detailed results for all 334 genes are summarized in Supplementary Tables 1 and 2. Of the 334 candidate genes examined in this study, 17 false positives were expected in each population due to chance alone. We observed exactly 17 genes with p-values ≤ 0.05 in the Danish sample by TRIMM and 20 genes by HAPLIN, while there were 24 and 27 such genes by the two methods in the Norwegian sample (Figure 1).
Table 1.
Sample distribution of isolated cleft lip only (CLO) and isolated cleft lip and cleft palate (CLP) by population and gender
| Population | Cleft type | Gender | Count |
|---|---|---|---|
| Norway | CLO | Male | 67 |
| Female | 54 | ||
| Norway | CLP | Male | 135 |
| Female | 55 | ||
| Denmark | CLO | Male | 47 |
| Female | 29 | ||
| Denmark | CLP | Male | 77 |
| Female | 30 |
Figure 1. Schweder-Spjøtvoll plotsof p-values.
TRIMM (A) and HAPLIN (B) analyses of the Norwegian and Danish isolated CLO samples. Genes with p-values ≤ 0.1 are shown on the X-axis and ordered according to their p-values (Y-axis). Those with p-values ≤ 0.05 are depicted in red. All genes with p-values ≤ 0.1 in both populations are indicated by vertical lines connecting the upper (Norway) and lower (Denmark) plots.
To differentiate between chance associations and those that are more likely linked with risk of isolated CLO, we looked for genes that showed a consistent pattern of association across both populations and analytic methods. Overall, TRIMM and HAPLIN both identified FGF12 among genes that were strongly associated with isolated CLO (with results replicated in both populations), but only TRIMM identified IRF6 and VCL (with replication in both populations). In contrast, only HAPLIN identified an association with CX43 in both populations. With TRIMM, CX43 was strongly identified in Denmark but just missed the criterion in Norway. When the distribution of the observed Fisher-combined p-values from the TRIMM and HAPLIN analyses was contrasted with the null distribution, three genes in particular—IRF6, FGF12 and CX43—showed marked deviations at the significant end of the distributions in the QQ plots (Figure 2).
Figure 2. Quantile-quantile (QQ) plots of p-values.
The QQ plot compares the distribution of the observed Fisher-combined p-values (−log10 scale) for both populations with an expected uniform distribution under the null (sloping line). The plots for isolated CLO are provided separately for TRIMM (A) and HAPLIN (B). Gene labels for the top six most significant genes are displayed in each plot.
Table 2 provides relative risk estimates and 95% confidence intervals for each candidate risk haplotype in each population. Because several of the risk haplotypes had low frequencies and the sample size of each population was relatively small, homozygotes were too rare to estimate R2 reliably (R2 being the relative risk parameter for offspring carrying two copies of the risk haplotype versus offspring carrying none). Hence, we enforced a log-additive risk structure to simplify the model and thereby approximate R2 as R12. With the exception of FGF12 in the Danish isolated CLO sample, the estimated proportion carrying one copy of the candidate risk haplotype was above 20%, allowing reliable estimation of R1. For the CX43 markers, opposite risk haplotypes were identified in Norway and Denmark, despite the two genotyped SNPs having very similar minor allele frequencies in the two populations. In the Danish sample, the relative risk of 5.49 was deemed implausible because of the wide confidence interval and the exceedingly low frequency of the FGF12 A-G-G candidate risk haplotype (2%). Overall, R1 ranged from 1.46 to 2.53 for fetuses inheriting one copy of the risk haplotype in the four genes tested.
Table 2.
Relative risk estimates for candidate risk haplotypes
| Gene | Sample | No. of SNPs | No. Tagging SNPs | Tagging SNPsa | Candidate risk haplotype | R1 (95% CI)b | Freq of 0 copy c | Freq of 1 copy c | Freq of 2 copies c |
|---|---|---|---|---|---|---|---|---|---|
| FGF12 | Norway | 6 | 3 | rs11717284, rs6790664, rs1464942 | A-C-T | 1.87 (1.23–2.84) | 0.71 | 0.27 | 0.02 |
| Denmark | 6 | 3 | rs1464942, rs12106855, rs1875735 | A-G-G | 5.49 (1.90–15.8) | 0.96 | 0.04 | 0.00 | |
|
| |||||||||
| IRF6 | Norway | 7 | 2 | rs680331, rs674433 | G-G | 1.81 (1.15–2.84) | 0.70 | 0.24 | 0.05 |
| Denmark | 7 | 3 | rs680331, rs674433, rs2013162 | G-G-C | 2.24 (1.28–3.65) | 0.62 | 0.35 | 0.02 | |
|
| |||||||||
| VCL | Norway | 5 | 1 | rs4746172 | C | 1.47 (1.01–2.12) | 0.53 | 0.42 | 0.06 |
| Denmark | 5 | 3 | rs10762573, rs2131960, rs4746172 | C-A-T | 1.46 (0.93–2.29) | 0.17 | 0.55 | 0.28 | |
|
| |||||||||
| CX43 | Norway | 2 | 2 | rs12197797, rs11961755 | C-G | 1.61 (1.07–2.43) | 0.09 | 0.47 | 0.45 |
| Denmark | 2 | 2 | rs12197797, rs11961755 | G-A | 2.53 (1.43–4.49) | 0.73 | 0.25 | 0.01 | |
χ2 test for deviation from Hardy-Weinberg equilibrium in the 592 Norwegian control triads showed p-values <0.05 for all these SNPs.
R1 is the relative risk for offspring heterozygous for the candidate risk haplotype relative to an offspring with no copies of that haplotype. We imposed a simplified log-additive risk model, where risk for offspring carrying two copies of the candidate risk haplotype is equivalent to R12. Thus, only one risk parameter, R1, is estimated for offspring effects.
Frequency of diplotypes with 0, 1 or 2 copies of the candidate risk haplotype.
Lastly, to verify that the above associations were specific for isolated CLO, we repeated the analyses on the larger samples of isolated CLP from the two populations. Remarkably, the associations with FGF12, IRF6, VCL and CX43 were no longer statistically significant (Supplementary Tables 3 and 4).
DISCUSSION
Given preliminary evidence that at least a subset of CLO may be etiologically distinct from CLP, and the growing realization that a more refined phenotype is paramount to the success of any gene-mapping effort (Jugessur et al., 2009a; Marazita, 2007; Marazita and Mooney, 2004; Morris et al., 2009; Weinberg et al., 2008; Weinberg et al., 2006), our objective was to evaluate fetal genetic risk of isolated CLO separately from isolated CLP. Two population-based samples of offspring-parent triads from Norway and Denmark were used to search for multi-marker transmission distortions among 334 autosomal candidate genes for orofacial clefts.
One way to assess replication in our data is to see whether more genes than expected by chance have p-values ≤ 0.1 in both Norway and Denmark. Given that 334 tests were performed, if those genes were all unlinked, one would expect about three genes (0.1 × 0.1 × 334) to “replicate” in this sense; i.e. having a line connecting them in Figure 1A or Figure 1B. For TRIMM, there are seven genes that replicate in this manner. For HAPLIN, there are 12 such genes—about nine more than expected. Overall, five genes (FGF12, IRF6, PVRL1, CX43 and VCL) appeared on both lists.
IRF6 and FGF12 were both found to be strongly associated with isolated CL/P in a recent analysis of the same dataset (Jugessur et al., 2009b). Of the large number of genes reported to be associated with isolated CL/P in the literature, IRF6 is by far the most consistent across studies. Mutations in IRF6 cause two allelic autosomal dominant clefting disorders, Van der Woude syndrome (VWS [MIM 119300]) and popliteal pterygium syndrome (PPS [MIM 119500]), which are characterized by varying degrees of cleft lip defects, lip pits, skin-folds, syndactyly and intra-oral epithelial adhesions (de Lima et al., 2009; Kondo et al., 2002). The observation that approximately15% of affected individuals are clinically indistinguishable from isolated clefts drove researchers to investigate whether variants in IRF6 might also be involved in the etiology of isolated clefts. This was subsequently demonstrated in multiple studies (reviewed in (Jugessur et al., 2008)). More recently, the identification of a common SNP (rs642961) that disrupted the binding site for transcription factor AP-2α within a highly conserved IRF6 enhancer element provided new insights into how IRF6 might influence the risk of isolated CL/P (Rahimov et al., 2008). That IRF6 and AP-2α are both implicated in clefting of the lip is all the more striking given an earlier report describing an essential role for AP-2α in cranial closure and craniofacial development (Schorle et al., 1996).
With particular relevance to the current analysis, Rahimov and co-workers found the frequency of the risk-conferring rs642961[A] allele to be significantly higher in cases with isolated CLO vs. other cleft phenotypes (Rahimov et al., 2008). There was a near doubling of the risk of isolated CLO with this allele compared with controls. This phenotype-specific risk for isolated CLO was further confirmed by our recent genome-scan linkage analysis of 820 multiplex CL/P families (Marazita et al., 2009), where the IRF6 region results were found to be most significant for families in which affected individuals had CLO alone.
The gene for poliovirus receptor-related 1 (PVRL1) encodes nectin-1 and is expressed primarily in the MEE of the palatal shelves, the ectodermal component of tooth buds, the olfactory epithelium and the skin surface epithelium (Suzuki et al., 2000). Homozygosity for a common nonsense mutation in this gene (W185X) results in an autosomal recessive clefting disorder known as CL/P-ectodermal dysplasia syndrome (CLPED1) (Suzuki et al., 2000). Heterozygosity for W185X has also been suggested to influence the risk of isolated CL/P (Sozen et al., 2001), with significant associations subsequently reported between genetic variants in PVRL1 and isolated CL/P (Avila et al., 2006; Neiswanger et al., 2006). Taken together, these findings indicate that both rare and common variants within PVRL1 may make minor contributions to the cleft lip phenotype.
In contrast with the above genes, FGF12, CX43 and VCL represent novel associations with isolated CLO. FGF12 encodes a member of the fibroblast growth factor (FGF) family of signaling molecules that are highly conserved evolutionarily and play important roles in craniofacial development (Nie et al., 2006). Several members of the FGF family and their receptors have previously been implicated in various human disorders, but FGF12 has not been studied. For example, mutations in FGFR1, 2, and 3 are associated with craniosynostosis and other facio-skeletal malformations (Pauws and Stanier, 2007). Diagnostic sequencing of 12 FGF-related genes in 184 individuals with isolated CL/P identified seven potential disease-causing mutations, including a nonsense mutation in FGFR1, a de novo missense mutation in FGF8, and other missense variants in FGFR1, FGFR2, and FGFR3 (Riley et al., 2007). FGF12 was also found to be associated with isolated CL/P in our previous analysis of the same dataset, in which no distinctions were made between CLO and CLP (Jugessur et al., 2009b). In this study, the association of FGF12 with isolated CLO and not with the larger sample of isolated CLP strongly suggests that the association with FGF12 is specific for isolated CLO, highlighting the importance of careful phenotypic delineation for more targeted gene-mapping.
Intriguingly, opposite directions of association were identified with the same risk haplotypes in Norway and Denmark for the connexin 43 gene [CX43, a.k.a. gap junction protein alpha 1 (GJA1)], although very similar minor allele frequencies were noted in both populations for the two genotyped SNPs. A false positive association is a possibility, but the apparent reversal of genetic effects across the two populations—referred to as a genetic “flip-flop” in the literature—does not necessarily mean a statistical artifact (Lin et al., 2007; Zaykin and Shibata, 2008). Multi-marker effects and variation in linkage disequilibrium in the Norwegian and Danish populations could have caused this apparently contradictory observation. Despite this inconsistency, the association with this gene is plausible on biological grounds. CX43 encodes a component of gap junctions that mediate the diffusion of ions and metabolites between the cytoplasm of adjacent cells. Mutations in CX43 cause both autosomal and recessive forms of oculodentodigital dysplasia [ODDD (MIM 164200)] that are characterized by developmental abnormalities of the face, eyes, limbs, and dentition (Paznekas et al., 2003). Anomalies in the craniofacial region include short palpebral fissures, mandible with wide alveolar ridge, hypoplastic nose, dental anomalies, and inverted palate. Whole-mount in situ hybridization shows strong expression of Gja1 in the murine frontonasal process, developing branchial arches, medial and lateral nasal processes, mandibular processes, and the shelves of the secondary palate at the time of fusion (Richardson et al., 2004). Consistent with the expression data, cleft palate has been reported in a proportion of ODDD patients. The strong expression of Gja1 at the fusion sites between the lateral nasal, medial nasal and maxillary prominences in E10.5 embryos points to a significant role for this gene in upper lip formation. To our knowledge, however, no associations with CX43 have previously been reported in orofacial clefting.
Vinculin (VCL) has a well-characterized function as a cytoskeletal protein, involved in the linking of actin filaments to integrins. This process enables cells to attach to each other and to the extracellular matrix. Overexpression of vinculin results in increased cell adhesion, whereas cells lacking vinculin are highly metastatic and motile (Ziegler et al., 2006). Vinculin is also implicated in modulating the signaling pathways involved in apoptosis (Subauste et al., 2005). The proper formation of the primary palate requires cells to adhere tightly to one another and to the extracellular matrix, and to undergo apoptosis at specific stages of development. As VCL is involved in all of these processes, it is plausible that aberrant function of this gene contributes to the pathogenesis of cleft lip. Like CX43, an association of VCL with isolated CLO has not been reported previously.
When performing a large number of tests, as in the present study, there is obviously a need to correct for multiple-testing. Both TRIMM and HAPLIN correct for within-gene multiple testing. HAPLIN uses 3 SNPs per sliding-window and calculates a score test p-value for each window, the smallest p-value is chosen. To adjust this p-value for within-gene multiple testing, we used the principle of “seemingly unrelated estimation” implemented in the Stata software. Thus, the individual score contributions for each sliding-window is saved and their between-window correlations is used to correct the p-value. TRIMM on the other hand achieves a natural correction for multiple testing by treating multiple SNP as a set and permuting alleles at all SNPs simultaneously to evaluate the test significance. Unfortunately, no single gene remained significant after the Bonferroni correction for the number of genes tested. However, it is generally agreed that the Bonferroni requirement of ensuring an overall type 1 error rate of below 5% is too strict and may result in too many false negatives (Rice TK, Schork NJ, Rao DC (2008)), particularly in this study where the selected genes already had an a priori connection to clefting. As an alternative, we focused here on genes showing significance in both populations.
Overall, a relatively small number of genes were found to be associated with isolated CLO among the 334 genes analyzed in this study, despite the particularly strong familial risk of recurrence for this group of birth defects (Sivertsen et al., 2008). This result may reflect the relatively small number of SNPs assayed per gene (range 1–12; mean 4), which may have provided only a partial representation of the genetic architecture of the candidate genes. All candidate genes in this study were selected a priori for their potential roles in orofacial clefting. There are almost definitely other candidate genes and genetic variations not included in the current selection (Birnbaum et al., 2009; Grant et al., 2009). It is also becoming increasingly clear that copy number variants due to small insertions and deletions may contribute to the risk of many human diseases (Cooper et al., 2008; Kidd et al., 2008). By studying apparent Mendelian inconsistencies between parents and offspring, we have previously confirmed deletions involving several genes, including both de novo and familial cases (Shi et al., 2009a). In particular, deletions of SUMO1, TBX1, and TFAP2A were predicted to be etiologic. Genetic effects might also operate through mechanisms that are unrelated to differences in linkage disequilibrium, as for example, in the presence of substantial allelic heterogeneity, where linkage disequilibrium is inoperative or would require impractical sample sizes to detect. Linkage analysis (used to identify FOXE1 for example (Moreno et al., 2009)) or eventually whole-exome or genome sequencing (Ng et al., 2009) will be required in this context for allele/gene discovery. Finally, interactions with environmental teratogens by genetic contributor may require a depth of exposure data not practical in current studies.
Although we had nearly 800 case-parent triads to begin with, our focus on isolated CLO (the smallest cleft subtype) resulted in a smaller sample size. We tried to maximize our chances of capturing an association signal by looking for replications across the two national samples. Reanalysis of the larger sample of isolated CLP triads did not replicate any of the significant associations detected in the isolated CLO triads, providing further support for the notion that at least a subset of CLO may have a different underlying cause than CLP. In addition, the strong associations observed with IRF6 and FGF12 in our previous analysis of isolated CL/P in the same dataset (Jugessur et al., 2009b) indicate that both TRIMM and HAPLIN are able to detect these CLO-specific association signals despite a mixture of the two cleft lip subtypes.
The custom designed panel of candidate genes remains to be a valuable resource for studying complex disease even after the widespread use of GWAS (Birnbaum et al. 2009; Grant et al. 2009; Mangold et al. 2010; Beaty et al. 2010). In addition to its cost-efficiency, the candidate gene based approach reduces the multiple testing problem. For a complex disease as clefts many genes may affect the risk with each contributing only an incremental amount. Correcting for the whole GWAS tests may kill any association signals that may nevertheless be revealed by using the custom designed panel of candidate genes.
In conclusion, our approach replicated previously reported associations of IRF6 and FGF12 with orofacial clefts, with evidence that these gene variants may act particularly on isolated CLO. In addition, we have identified new associations of VCL and CX43 with orofacial clefts—specifically with CLO — and have replicated these associations in two independent populations. The strength of these associations and the biological plausibility of these genes in clefting of the lip warrant further studies in other isolated CLO populations.
Supplementary Material
Acknowledgments
Grant information: This research was supported in part by the Intramural Research Program of the US National Institute of Environmental Health Sciences (NIEHS); by US National Institutes of Health (NIH) grants DE08559, P60 DE13076, NIH P30 ES05605, and RO1 DE-11948-04; and by the Norwegian Research Council (NFR 177522).
We thank all participating families who made this study possible. Special thanks go to the following individuals for their input in candidate gene selection and sample preparation: Kathy Frees, Ecaterina Dragan, Adela Mansilla, Bridget Riley, Dorthe Grosen, Brian Schutte, Edward Lammer, Temis Felix, Michael Lovett, Satoshi Suzuki, Andrew Olshan, Rulang Jiang, Diana Caprau, Richard Finnell, Christine Jewell, John Cidlowski, Fedik Rahimov, Jack Taylor and Alexandre Vieira. We are particularly grateful to Susie McConnell and Jacqueline Leung-Heras for outstanding secretarial support. Genotyping services were provided by the Center for Inherited Disease Research (CIDR), which is funded through a federal contract from the National Institutes of Health (NIH) to the Johns Hopkins University (grant N01-HG-65403). We thank Ivy McMullen, Corinne Boehm, Kim Doheny, and other CIDR staff involved in this project. We also thank the US National Institute of Dental and Craniofacial Research (NIDCR) for underwriting a significant proportion of the genotyping costs by CIDR.
Footnotes
Competing Interest: None.
References
- Avila JR, Jezewski PA, Vieira AR, Orioli IM, Castilla EE, Christensen K, Daack-Hirsch S, Romitti PA, Murray JC. PVRL1 variants contribute to non-syndromic cleft lip and palate in multiple populations. American journal of medical genetics. 2006;140(23):2562–2570. doi: 10.1002/ajmg.a.31367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille C, Knudsen LB, Christensen K. Changing lifestyles and oral clefts occurrence in Denmark. Cleft Palate Craniofac J. 2005;42(3):255–259. doi: 10.1597/03-139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, Mangold E. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature genetics. 2009;41(4):473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nature genetics. 2008;40(10):1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima RL, Hoper SA, Ghassibe M, Cooper ME, Rorick NK, Kondo S, Katz L, Marazita ML, Compton J, Bale S, Hehr U, Dixon MJ, Daack-Hirsch S, Boute O, Bayet B, Revencu N, Verellen-Dumoulin C, Vikkula M, Richieri-Costa A, Moretti-Ferreira D, Murray JC, Schutte BC. Prevalence and nonrandom distribution of exonic mutations in interferon regulatory factor 6 in 307 families with Van der Woude syndrome and 37 families with popliteal pterygium syndrome. Genet Med. 2009;11(4):241–247. doi: 10.1097/GIM.0b013e318197a49a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. Statistical Methods for Research Workers. New York: Hafner; 1958. [Google Scholar]
- Genisca AE, Frias JL, Broussard CS, Honein MA, Lammer EJ, Moore CA, Shaw GM, Murray JC, Yang W, Rasmussen SA. Orofacial clefts in the National Birth Defects Prevention Study, 1997–2004. American journal of medical genetics. 2009;149A(6):1149–1158. doi: 10.1002/ajmg.a.32854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjessing HK, Lie RT. Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes. Annals of human genetics. 2006;(69):1–15. doi: 10.1111/j.1529-8817.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, Frackelton EC, Otieno FG, Chiavacci RM, Nah HD, Kirschner RE, Hakonarson H. A Genome-Wide Association Study Identifies a Locus for Nonsyndromic Cleft Lip with or without Cleft Palate on 8q24. J Pediatr. 2009 doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Grosen D, Chevrier C, Skytthe A, Bille C, Molsted K, Sivertsen A, Murray JC, Christensen K. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. J Med Genet. 2009 doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harville EW, Wilcox AJ, Lie RT, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? American journal of epidemiology. 2005;162(5):448–453. doi: 10.1093/aje/kwi214. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral diseases. 2009a;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Rahimov F, Lie RT, Wilcox AJ, Gjessing HK, Nilsen RM, Nguyen TT, Murray JC. Genetic variants in IRF6 and the risk of facial clefts: single-marker and haplotype-based analyses in a population-based case-control study of facial clefts in Norway. Genetic epidemiology. 2008;32(5):413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, Christensen K, Boyles AL, Daack-Hirsch S, Trung TN, Bille C, Lidral AC, Murray JC. Genetic determinants of facial clefting: analysis of 357 candidate genes using two national cleft studies from Scandinavia. PLoS ONE. 2009b;4(4):e5385. doi: 10.1371/journal.pone.0005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nature genetics. 2002;32(2):285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. American journal of human genetics. 2007;80(3):531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML. Segregation analyses. In: Wyszynski DF, editor. Cleft Lip and Palate: From Origin to Treatment. New York: Oxford University Press; 2002. pp. 222–233. [Google Scholar]
- Marazita ML. Subclinical features in non-syndromic cleft lip with or without cleft palate (CL/P): review of the evidence that subepithelial orbicularis oris muscle defects are part of an expanded phenotype for CL/P. Orthodontics & craniofacial research. 2007;10(2):82–87. doi: 10.1111/j.1601-6343.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, Cooper ME, Govil M, Daack-Hirsch S, Riley B, Jugessur A, Felix T, Moreno L, Mansilla MA, Vieira AR, Doheny K, Pugh E, Valencia-Ramirez C, Arcos-Burgos M. Genome Scan, Fine-Mapping, and Candidate Gene Analysis of Non-Syndromic Cleft Lip with or without Cleft Palate Reveals Phenotype-Specific Differences in Linkage and Association Results. Hum Hered. 2009;68(3):151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Mooney MP. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin Plast Surg. 2004;31(2):125–140. doi: 10.1016/S0094-1298(03)00138-X. [DOI] [PubMed] [Google Scholar]
- Mitchell LE, Beaty TH, Lidral AC, Munger RG, Murray JC, Saal HM, Wyszynski DF. Guidelines for the design and analysis of studies on nonsyndromic cleft lip and cleft palate in humans: summary report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate Craniofac J. 2002;39(1):93–100. doi: 10.1597/1545-1569_2002_039_0093_gftdaa_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, Johnson MK, Brauer D, Krahn K, Daack-Hirsch S, L’Heureux J, Valencia-Ramirez C, Rivera D, Lopez AM, Moreno MA, Hing A, Lammer EJ, Jones M, Christensen K, Lie RT, Jugessur A, Wilcox AJ, Chines P, Pugh E, Doheny K, Arcos-Burgos M, Marazita ML, Murray JC, Lidral AC. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Human molecular genetics. 2009;18(24):4879–4896. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Lindgren CM, Zeggini E, Timpson NJ, Frayling TM, Hattersley AT, McCarthy MI. A powerful approach to sub-phenotype analysis in population-based genetic association studies. Genet Epidemiol. 2009 doi: 10.1002/gepi.20486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski DFE, editor. Cleft lip and palate: from origin to treatment. New York: Oxford University Press; 2002. [Google Scholar]
- Neiswanger K, Deleyiannis FW, Avila JR, Cooper ME, Brandon CA, Vieira AR, Noorchashm N, Weinberg SM, Bardi KM, Murray JC, Marazita ML. Candidate genes for oral-facial clefts in Guatemalan families. Ann Plast Surg. 2006;56(5):518–521. doi: 10.1097/01.sap.0000210261.65455.9d. discussion 521. [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. 2006;12(2):102–111. doi: 10.1111/j.1601-0825.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E, Stanier P. FGF signalling and SUMO modification: new players in the aetiology of cleft lip and/or palate. Trends Genet. 2007;23(12):631–640. doi: 10.1016/j.tig.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72(2):408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2006. [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R, Donnai D, Meire F, Dixon MJ. Expression of Gja1 correlates with the phenotype observed in oculodentodigital syndrome/type III syndactyly. J Med Genet. 2004;41(1):60–67. doi: 10.1136/jmg.2003.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M, Marazita ML, Murray JC. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007;104(11):4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381(6579):235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Schweder T, Spjøtvoll E. Plots of P-values to evaluate many tests simultaneously. Biometrika. 1982;69(3):493–502. [Google Scholar]
- Shi M, Mostowska A, Jugessur A, Johnson MK, Mansilla MA, Christensen K, Lie RT, Wilcox AJ, Murray JC. Identification of microdeletions in candidate genes for cleft lip and/or palate. Birth Defects Res A Clin Mol Teratol. 2009a;85(1):42–51. doi: 10.1002/bdra.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Umbach DM, Weinberg CR. Identification of Risk-Related Haplotypes with the Use of Multiple SNPs from Nuclear Families. American journal of human genetics. 2007;81(1):53–66. doi: 10.1086/518670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Umbach DM, Weinberg CR. Using Case-parent Triads to Estimate Relative Risks Associated with a Candidate Haplotype. Ann Hum Genet. 2009b;73(Pt 3):346–359. doi: 10.1111/j.1469-1809.2009.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsen A, Wilcox AJ, Skjaerven R, Vindenes HA, Abyholm F, Harville E, Lie RT. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ (Clinical research ed. 2008;336(7641):432–434. doi: 10.1136/bmj.39458.563611.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen MA, Suzuki K, Tolarova MM, Bustos T, Fernandez Iglesias JE, Spritz RA. Mutation of PVRL1 is associated with sporadic, non-syndromic cleft lip/palate in northern Venezuela. Nat Genet. 2001;29(2):141–142. doi: 10.1038/ng740. [DOI] [PubMed] [Google Scholar]
- Subauste MC, Nalbant P, Adamson ED, Hahn KM. Vinculin controls PTEN protein level by maintaining the interaction of the adherens junction protein beta-catenin with the scaffolding protein MAGI-2. J Biol Chem. 2005;280(7):5676–5681. doi: 10.1074/jbc.M405561200. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helms JA, Spritz RA. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 2000;25(4):427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Brandon CA, McHenry TH, Neiswanger K, Deleyiannis FW, de Salamanca JE, Castilla EE, Czeizel AE, Vieira AR, Marazita ML. Rethinking isolated cleft palate: evidence of occult lip defects in a subset of cases. American journal of medical genetics. 2008;146A(13):1670–1675. doi: 10.1002/ajmg.a.32291. [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Neiswanger K, Martin RA, Mooney MP, Kane AA, Wenger SL, Losee J, Deleyiannis F, Ma L, De Salamanca JE, Czeizel AE, Marazita ML. The Pittsburgh Oral-Facial Cleft study: expanding the cleft phenotype. Background and justification. Cleft Palate Craniofac J. 2006;43(1):7–20. doi: 10.1597/04-122r1.1. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Lie RT, Solvoll K, Taylor J, McConnaughey DR, Abyholm F, Vindenes H, Vollset SE, Drevon CA. Folic acid supplements and risk of facial clefts: national population based case-control study. BMJ (Clinical research ed. 2007;334(7591):464. doi: 10.1136/bmj.39079.618287.0B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Shibata K. Genetic flip-flop without an accompanying change in linkage disequilibrium. American journal of human genetics. 2008;82(3):794–796. doi: 10.1016/j.ajhg.2008.02.001. author reply 796–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16(9):453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.