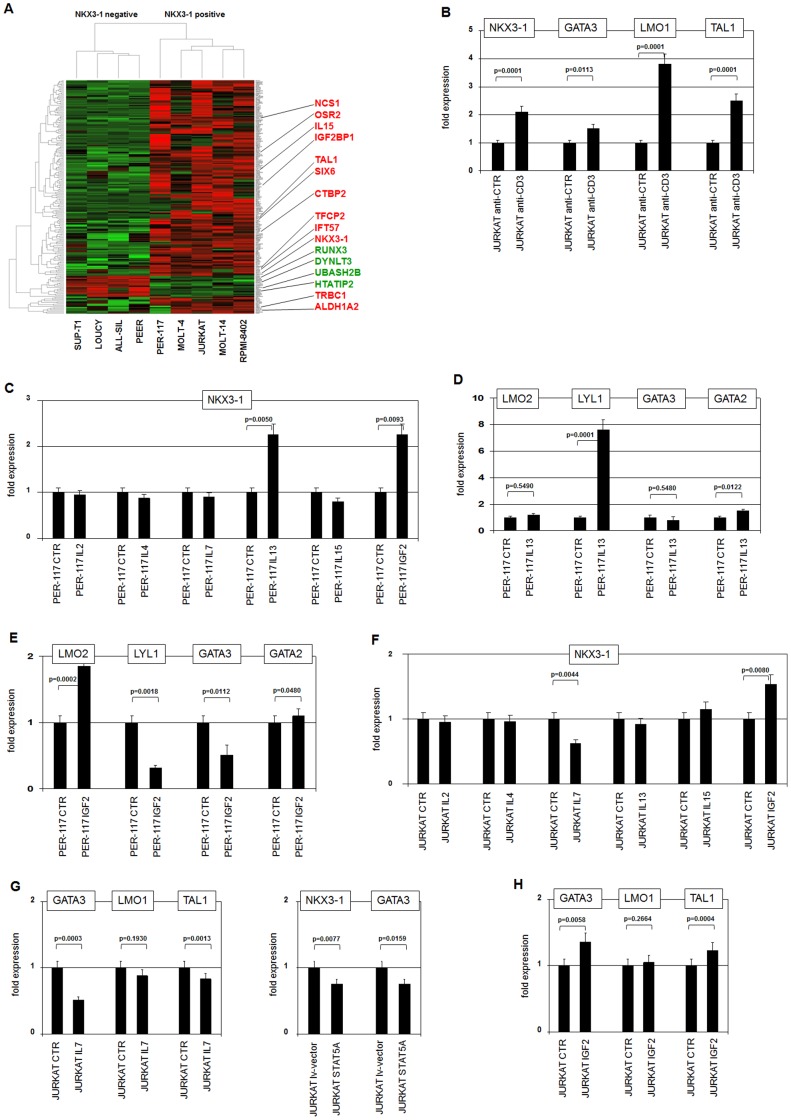
Figure 4. Identification of additional NKX3-1 regulators.
(A) Heat map of 209 genes which displayed a significant differential expression between two groups of T-ALL cell lines concerning NKX3-1 expression, as analysed by the LIMMA package. Four T-ALL cell lines (SUP-T1, LOUCY, ALL-SIL, PEER) are NKX3-1 negative while five others (PER-117, MOLT-4, JURKAT, MOLT-14, RPMI-8402) are NKX3-1 positive. Red indicates strong expression levels, green low levels and black medium levels. Sixteen conspicuously expressed genes are highlighted. (B) RQ-PCR analysis of JURKAT cells treated with activating anti-CD3 antibody demonstrates concomitantly upregulation of NKX3-1, GATA3, LMO1 and TAL1. (C) Treatment of PER-117 cells with specific ligands demonstrates increased expression of NKX3-1 after stimulation with IL13 or IGF2. (D) Treatment of PER-117 cells with IL13 resulted in strong activation of LYL1 and slight activation of GATA2 expression, while that of LMO2 and GATA3 remained unperturbed. (E) Treatment of PER-117 cells with IGF2 resulted in differentially altered expression levels of LMO2, LYL1 and GATA3 while GATA2 remained unchanged. (F) Treatment of JURKAT cells with specific ligands demonstrates decreased expression of NKX3-1 after stimulation with IL7, while the expression increased after stimulation with IGF2. (G) Treatment of JURKAT cells with IL7 resulted in decreased expression of GATA3, LMO1 and TAL1 (left). Lentiviral overexpressed STAT5A in JURKAT cells resulted in decreased expression of both NKX3-1 and GATA3 (right). (H) Treatment of JURKAT cells with IGF2 resulted in marginally increased expression of GATA3 and TAL1 while no change was visible for LMO1.