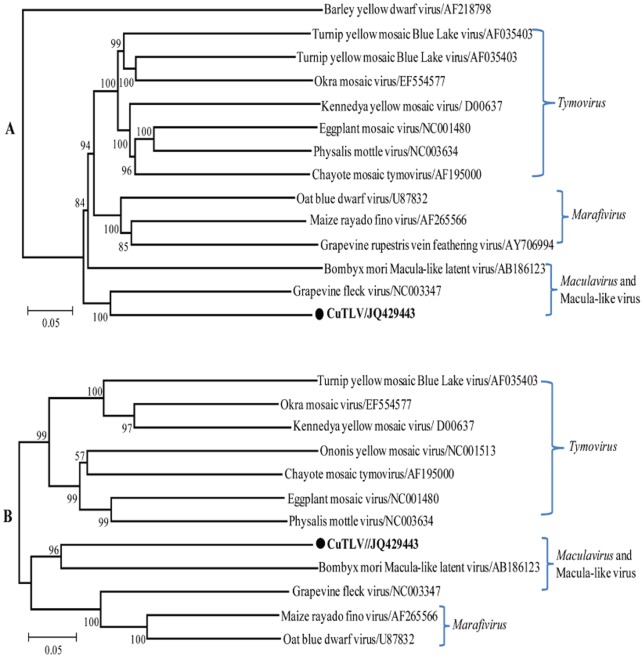
Figure 4. Phylogenetic tree of CuTLV based on the RP (A) and CP (B) sequences.
Phylogenetic analyses were performed by the neighbor-joining method using MEGA version 4 (www.megasoftware.net). Bootstrap probabilities of each node were calculated with 1000 replicates. The RP (A) tree was rooted by using barley yellow dwarf virus sequence as the out group virus. For CP (B) tree, no outgroup was included. Horizontal branch lengths are proportional to genetic distance and vertical branch lengths have no significance. The scale indicates the number of nucleotide substitutions per site. Sequence of CuTLV from this study is in boldface.