Abstract
In the dog, previous analyses of major histocompatibility complex (MHC) class I genes suggest a single polymorphic locus, Dog Leukocyte Antigen (DLA)-88. While 51 alleles have been reported, estimates of prevalence have not been made. We hypothesized that, within a breed, DLA-88 diversity would be restricted, and one or more dominant alleles could be identified. Accordingly, we determined allele usage in 47 Golden Retrievers and 39 Boxers. In each population, 10 alleles were found; 4 were shared. Seven novel alleles were identified. DLA-88*05101 and *50801 predominated in Golden Retrievers, while most Boxers carried *03401. In these breeds DLA-88 polymorphisms are limited and largely non-overlapping. The finding of highly prevalent alleles fulfills an important prerequisite for studying canine CD8+ T-cell responses.
Keywords: breeds, canine, class I genes, major histocompatibility complex, polymorphisms
Classical major histocompatibility complex (MHC) class I molecules are widely expressed, cell surface glycoproteins that regulate the development and function of CD8+ T-cell responses through the presentation of cytosolic-origin peptide epitopes. In part, these class I molecules are distinguished from their nonclassical counterparts by extensive polymorphisms. In humans, for example, there are >1500 alleles at the Human Leukocyte Antigen (HLA)-A locus, >2000 alleles at the HLA-B locus, and >1000 alleles at the HLA-C locus (1). Allelic variation, which arises primarily by intra-locus recombination or point mutation, is frequently concentrated in hypervariable regions (HVRs), where residues of the heavy chain α1 and α2 domains contact the bound peptide (2). Such variation can have important consequences. In a murine model, an H-2K allele with differences only in the peptide-binding determinants was associated with increased diversity of the corresponding cytotoxic T-cell repertoire and enhanced resistance to viral infection (3). Selection pressure from pathogens can act on allelic variation to maintain the polymorphism of classical MHC molecules (4). Not surprisingly, restricted MHC diversity can have devastating effects on population survival, as illustrated by the particularly dramatic example of transmissible facial tumors of Tasmanian devils (5).
In the domestic dog, seven class I loci have been identified in the Dog Leukocyte Antigen (DLA) complex, and of these, four genes are transcribed: DLA-12, -64, -88 (all on chromosome 12) and -79 (on chromosome 18) (6, 7). The DLA-88 locus appears to be the most polymorphic, with 51 published alleles identified from >205 unrelated dogs (8-11). Variability at the other loci appears much more limited, with two, three and four alleles described to date for DLA-12, -64 and -79, respectively, from samples obtained from 18-20 dogs (8). While there have been no large scale DLA class I sequencing efforts published so far, it is reasonable to estimate that the overall pool of diversity in dogs will be much smaller than in humans, given the potentially low number of founders, and the recency of domestication, which is too short an evolutionary time to permit the creation of new alleles in any significant number (12). With some estimations of as few as 50 to 100 founding wolves (13), the total number of class I alleles at the DLA-88 locus may be no greater than several hundred in the domesticated dog population. This supposition is consistent with class II data, with a total of only 100 reported alleles at the most variable locus, DLA-DRB1, obtained from >1600 dogs (14).
The polymorphisms of MHC molecules constitute a barrier to allotransplantation, and also represent a substantial obstacle to studying antigen-specific T-cell responses across unrelated individuals. Advances in understanding cytotoxic CD8+ T-cell activity in autoimmune, infectious, and neoplastic canine diseases, as well as with immunization or transplantation, could be expedited by defining frequently occurring class I alleles in subpopulations of dogs. Breeds are an obvious choice for performing such analyses, as one would expect allelic diversity to be low, due to the limited number of foundation stock, as well as the use of inbreeding and overutilization of popular sires. Moreover, the easily recognized physical characteristics that define breed members could be a convenient means of identifying dogs that have a potentially high likelihood of sharing alleles or haplotypes. Indeed, intrabreed variation at several canine class II loci is quite limited (15-17). To date, however, no assessment of the prevalence of canine MHC class I alleles has been performed, while in humans, such information is readily available (18). In this investigation, we hypothesized that, within a breed, DLA-88 polymorphisms would be restricted, and one or a few alleles would dominate the locus. To examine this prediction, we sought to determine allelic variation in cohorts of dogs from two popular breeds, Golden Retrievers and Boxers, using reverse transcription-polymerase chain reaction (RT-PCR) and sequence-based typing.
For the study, venous blood samples were obtained from unrelated adult dogs (n = 65) admitted to the North Carolina State University (NCSU) Veterinary Teaching Hospital (VTH), from volunteer donors (n = 24) recruited through the Clinical Studies Core of the NCSU Center for Comparative Medicine and Translational Medicine, or from Beagles (n = 2) from the NCSU Laboratory Animal Resources unit. The blood collection protocol was approved by the NCSU Institutional Animal Care and Use Committee. Peripheral blood mononuclear cells (PBMCs) were isolated by centrifugation (400 g × 30 min at room temperature) over a Histopaque 1.077 continuous density gradient (Sigma-Aldrich, St. Louis, MO). From lysates of PBMCs, or the canine histiocytic cell line DH82 (ATCC CRL-10389), RNA was purified using an RNeasy Plus Mini Kit (Qiagen, Valencia, CA), and cDNA was synthesized by RT using an oligo(dT)15 primer.
Primers for the DLA-88 gene were synthesized by Invitrogen (Carlsbad, CA): exon 1 forward 5′-CGGAGATGGAGGTGGTGA-3′ and exon 4 reverse 5′-GGTGGCGGGTCACACG-3′. Amplification of cDNA templates was performed using a high fidelity Advantage-HF 2 PCR Kit (Clontech, Mountain View, CA) on a Mastercycler Pro thermocycler (Eppendorf, Hauppauge, NY) programmed with the following cycling conditions: initial denaturation/hot start at 95°C for 3 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 62°C for 30 s, extension at 68°C for 1 min and 15 s, succeeded by a final elongation step at 68°C for 5 min. All experiments included a negative control sample in which water was substituted for cDNA. Reaction products were separated by electrophoresis on a 1% agarose gel, excised, purified, and ligated overnight at 4° C into a pGEM-T Easy Vector (Promega, Madison, WI). Transformed colonies of chemically competent E. coli (GC10; Genesee Scientific, San Diego, CA) were selected by blue/white screening, and plasmids were purified by alkaline lysis (QIAprep Spin Miniprep Kit; Qiagen). Plasmids positive by EcoRI restriction digest were sequenced in both directions by Eurofins MWG Operon (Huntsville, AL) or Eton Bioscience Inc. (San Diego, CA), using T7 and SP6 primers. The obtained sequences were compared to all previously reported alleles of DLA-12,-64, -79, and -88 (8-11) using A plasmid Editor (ApE) software (19). For dogs with a single allele, a minimum of six colonies were sequenced; for heterozygotes, a minimum of two colonies were sequenced, if the alleles had been described previously. Sequences of novel alleles were determined from a minimum of six colonies, which were obtained from two or more different animals whenever possible, to insure that the observed allele was not due to PCR or sequencing error. Names for new alleles were assigned according to published conventions of the Comparative MHC Nomenclature Committee (20) by the curator (LJ Kennedy) of the canine IPD-MHC database.
We designed primers to anneal to exons 1 and 4 of the DLA class I heavy chain to provide full-length sequence data for exons 2 and 3, which encode the α1 and α2 domains that contain the antigen recognition site (ARS) for the T cell receptor. In genotyping HLA-A, B and C alleles, nucleotide sequences from exons 2 and 3 can unambiguously assign allelic identity in virtually all (>99.9%) cases (2), and accordingly, we used this approach in our study. Because of the high degree of homology between exons of the four transcribed MHC class I loci, it was not possible to produce individually specific forward and reverse primers for DLA-88 amplification from cDNA. To circumvent this difficulty, the forward primer was designed to anneal to the DLA-64 and -88 loci, while the reverse primer was designed to anneal to the DLA-12 and -88 loci. To confirm the ability of the primer pair to exclusively amplify transcripts from the DLA-88 locus, we first tested cDNA from the canine cell line DH82, from which only a single DLA-88 allele had been found previously (21); although unnamed, our analysis suggested that this sequence most likely represented DLA-88*50801 (with 2 bp substitutions). Cloning of the ∼650 bp amplimer yielded two sequences: DLA-88*50801 and a putative new allele (now DLA-88*05101). Because the locus of origin of the second allele could not be definitively established by reference to DLA-88 sequences in the canine IPD-MHC database, however, we repeated our evaluation of the primer set using cDNA from PBMCs isolated from a Beagle. As most reported alleles of DLA-88 have been obtained from Beagles or Beagle mixes (9, 11, 22), the likelihood of identifying another novel DLA-88 allele was presumed to be low. Indeed, only the previously reported allele DLA-88*50201 was sequenced in 23 colonies from this dog, indicating that amplification from the other class I loci – DLA-12,-64, and -79 – was highly unlikely with our set of primers. To provide further support for this conclusion, we then amplified cDNAs from four additional Beagles; again, only established DLA-88 alleles were identified in these analyses (DLA-88*00401, *00501, *00601, *50101, *50201).
We then sought to determine whether frequently shared alleles of DLA-88 could be identified within a breed. To accomplish this objective, we compared nucleotide sequences for exons 2 and 3 from 47 Golden Retrievers. From this population, a total of 10 alleles were obtained; the relative frequencies are shown in Table 1. Four novel alleles – DLA-88*01202,*02101,*03301 and *05101 – were found (Table 2). The most prevalent allele was DLA-88*50801. Together with DLA-88*05101, these two variants constituted 58% of the total allelic pool. When the frequency of occurrence was calculated on a per-dog basis, DLA-88*05101 was the most commonly encountered allele (Table 1, right column); the difference in prevalence between these two measures is attributable to the greater number of homozygotes among dogs possessing DLA-88*50801 (n = 11) than those with DLA-88*05101 (n = 4). Of the Golden Retrievers carrying the remaining eight alleles, only four homozygotes were identified. Not surprisingly, the DH82 cell line, which was derived from a Golden Retriever in Ohio in the mid-1980s (23), possessed the two most common alleles of the breed.
Table 1. Prevalence of DLA-88 alleles by breed.
| Golden Retriever (n = 47) | ||
|---|---|---|
|
| ||
| Allele | Allelic Frequencya | Phenotypic Frequencyb |
| *00201 | 1.1 | 2.2 |
| *00501 | 9.6 | 17.4 |
| *00601 | 1.1 | 2.2 |
| *01202 | 5.3 | 8.7 |
| *02101 | 6.4 | 13 |
| *03301 | 2.1 | 4.4 |
| *03801 | 13.8 | 26.1 |
| *05101 | 27.7 | 47.8 |
| *50101 | 1.1 | 2.2 |
| *50801 | 31.9 | 40.4 |
|
| ||
| Boxer (n = 39) | ||
|
| ||
| Allele | Allelic Frequency | Phenotypic Frequency |
|
| ||
| *00501 | 1.2 | 2.6 |
| *01201 | 4.7 | 10.3 |
| *02801 | 1.2 | 2.6 |
| *02803 | 12.9 | 34.4 |
| *02901 | 12.9 | 34.4 |
| *03201 | 2.4 | 2.6 |
| *03401 | 50.6 | 82.1 |
| *05101 | 2.4 | 5.1 |
| *50101 | 2.4 | 2.6 |
| *50801 | 9.4 | 17.9 |
Alleles common to both breeds are shown in bold type.
Numbers indicate percentage. Homozygote dogs were considered to possess two copies of the allele.
The percentage of dogs with the indicated allele.
Table 2. Novel DLA-88 alleles in the study populations.
| Allele | Number of clones | Number of dogs | Breeda | GenBank accession |
|---|---|---|---|---|
| *05101 | 101 | 24 | B; GR | HQ340121 |
| *02803 | 29 | 11 | B | HQ340113 |
| *02901 | 23 | 11 | B | HQ340112 |
| *02101 | 18 | 6 | GR | HQ340114 |
| *01202 | 17 | 4 | GR | HQ340115 |
| *03301 | 8 | 2 | GR | HQ340117 |
| *03201 | 8 | 1 | B | HQ340116 |
B, Boxer; GR, Golden Retriever
To demonstrate that the finding of a few dominant class I alleles was not peculiar to Golden Retrievers, we performed the same analysis with another breed, the Boxer. Among these dogs (n = 39), 10 DLA-88 alleles were again found; of these, only 4 were also common to Golden Retrievers (Table 1, bold face alleles). DLA-88*03401 constituted 50% of alleles in the total pool (Table 1, middle column) and was present in 82% of Boxers (right column). Seven dogs were homozygous for DLA-88*03401, while for all other nine alleles, only seven homozygote animals were identified. Four of the alleles found in Boxers are new: DLA-88*02803,*02901,*03201 and *05101 (Table 2).
Interestingly, during our analysis of Boxers, we observed that one dog appeared to have three DLA-88 alleles. To rule out contamination as a possible source of this finding, RNA was isolated from a second blood sample from this dog; genotyping again revealed the same three alleles. Ultimately, 7 of the 39 Boxers (5 males, 2 females) were observed to possess three different alleles. To our knowledge, none of these dogs had received a blood transfusion. Maternal microchimerism has been documented to result in the recovery of non-inherited HLA alleles from the PBMCs of healthy adults (24); however, this seems an unlikely cause, given the phenomenon was confined to Boxer dogs, and all of the individuals had the identical triplet combination: DLA-88*02803, *02901 and *03401 (note: in subsequent work, we have observed a few other combinations, but these always have contained either DLA-88*02803 or *02901 or both [Ross P, unpublished observations]). Therefore, we also considered the possibility that our primer pair had amplified sequences from other class I loci. Given that DLA-12, -64 and -79 are minimally polymorphic, one would predict such off-target amplification to occur in a majority of the dogs, however, not simply in a small subset of one breed. Moreover, sequence analysis strongly supports DLA-88 as the locus of origin for the two novel alleles, *02803 and *02901 (*03401 has been previously established), as pairwise identity of exons 2 and 3 of either allele with DLA-88*00101 is 97%, but only 82% with DLA-64 and 76% with DLA-79. The DLA-12 and -88 loci have far greater homology of exons 2 and 3, making this comparison much less useful; however, exon 1 sequences (encoding the leader peptide) are substantially disparate (6). Accordingly, we analyzed exon 1 sequences from *02803 and *02901, and found 100% identity with DLA-88. Nor do any of the three alleles appear to be pseudogenes, as all had open reading frames when sequenced through the transmembrane domain (data not shown). It should be stated that other investigators also have found three DLA-88 alleles specifically in dogs of the Boxer breed (LJ Kennedy, personal communication, University of Manchester, Manchester, UK). Additionally, dogs possessing three DQB1 alleles (with an analogous breed bias - Samoyeds) have been reported (14). For both loci – DLA-88 and -DQB1 – it is likely that this phenomenon is due to gene duplication, which has been found in the MHC of other species, such as the horse and cow (25, 26).
The alignment of the predicted amino acid sequences of the α1 and α2 domains of the seven novel alleles identified in Golden Retrievers and Boxers is shown in Figure 1. As expected, most of the amino acid variability is found in the defined HVRs (2). Outside of the HVRs, all amino acid differences identified in the new sequences, with the exceptions of pro 43, arg 50, glu 61 and glu 62 in DLA-88*05101, have been observed in at least one other allele. Of the 19 α1 and α2 residues in the β sheet of HLA-A2 that interact with β-2 microglobulin (27), sixteen are conserved across these canine alleles (Supplemental Figure 1). Similarly, all nine amino acids in the peptide binding region that are conserved in human and mouse classical class I molecules (27) are present, as are the cysteine residues that form the disulfide linkage between the α2 helix (cys 164) and the floor of the binding groove (cys 101) (28). Additionally, all of these new alleles have exon 1 sequences that align with the DLA-88 locus (data not shown). It is also worthy of note that, during this investigation, three additional sequences that were putative DLA-88 alleles were found (Supplemental Table 1); however, we were unable to obtain a sufficient number of colonies to validate these sequences.
Figure 1.
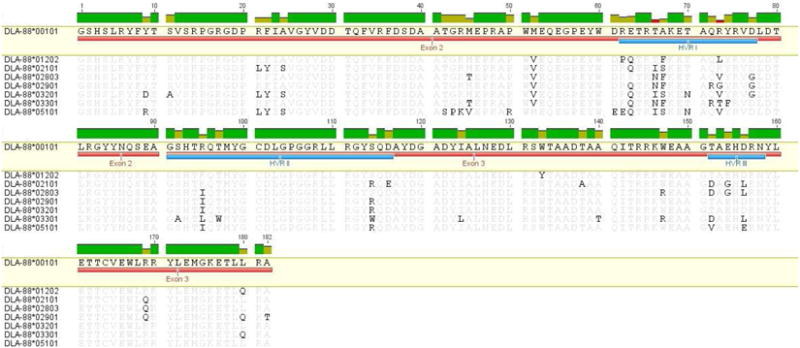
Alignment of the predicted amino acid sequences of exons 2 and 3 (marked by red bars) for seven novel DLA-88 alleles found in the study populations. Residues in light gray indicate identities with the reference sequence DLA-88*00101 (top), while those in dark gray indicate differences. The three HVRs are shown by the blue bars. For the naming of alleles, sequences with nucleotide differences that resulted in amino acid substitutions within any of the HVRs were designated as a new major type, (indicated by the first three digits), while sequences in which substitutions were confined to areas outside the HVRs were considered novel subtypes and numbered accordingly (last two digits).
To investigate the relationship of the new alleles with those that have been previously reported (8-11), we created a phylogenetic tree of all nucleotide sequences (Figure 2), analogous to such analyses of class I alleles in other species (29-31). As expected, the new alleles (marked by arrows) followed branch points that led to tight clustering with other known DLA-88 sequences. Further, these alleles are interspersed throughout the tree, consistent with the known creation histories for these relatively young (mid-to-late 19th century) breeds from disparate founders (wavy-coated Retriever, Tweed Water Spaniel, Irish setter, Bloodhound, and the St. John's Water Dog for the Golden Retriever; the Bullenbeisser and the English bulldog for the Boxer) (32, 33).
Figure 2.
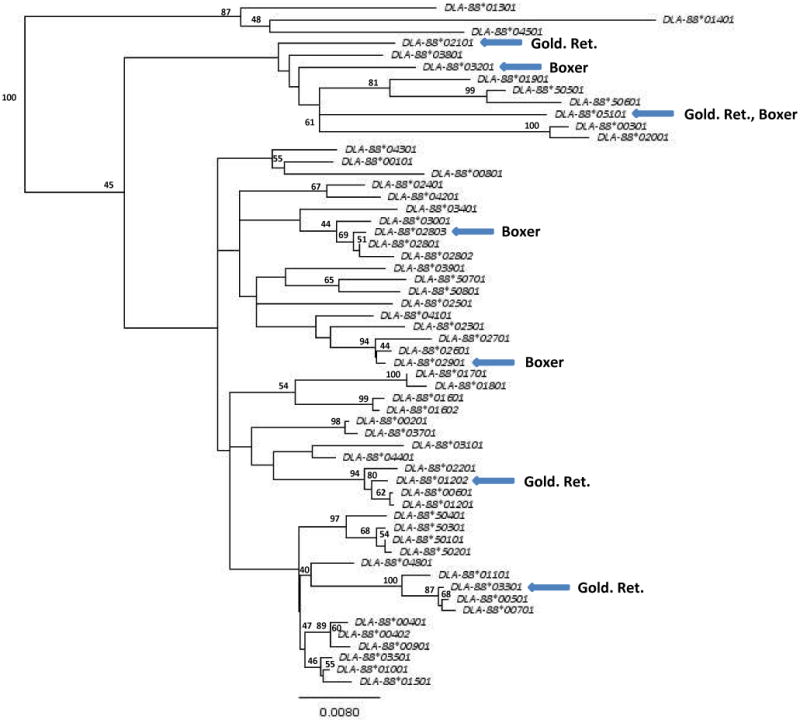
Phylogram of relationships between all known DLA-88 alleles. The seven new alleles identified in Golden Retrievers (GR) and Boxers (B) are indicated by the arrows. The tree was constructed with Geneious v.5.1 (52) from the concatenated nucleotide sequences of exons 2 and 3 on the basis of Tamura-Nei genetic distances (53), using the neighbor-joining method (54), and was rooted by the inclusion of HLA-A*0101 (GenBank AJ278305) as an outgroup. The numbers to the left of the internodes are the percentage of supporting bootstrap (n = 1000) replications; for clarity, only values ≥40 are shown. The units for the scale bar are the number of nucleotide substitutions per site.
Across species, a common characteristic of classical MHC class I molecules is the finding of positive selection of amino acid sites in the ARS (30, 31, 34, 35). We therefore evaluated all 58 DLA-88 nucleotide sequences – the 7 alleles from this study and the 51 alleles previously described – for evidence of positive selection using Bayesian analysis. Five codons in the α1/α2 domains with a mean probability >95% of being positively selected were identified: 73, 95, 114, 152 and 156. To assess the potential biologic relevance of these findings, we used the crystal structure of HLA-A*1101 to generate a 3-D model of one highly prevalent canine allele, DLA-88*03401, which is depicted in Figure 3. From this modeling, it can be seen that all of the positively selected sites are predicted to occur in the ARS (specifically, the peptide binding groove) of this class I molecule.
Figure 3.
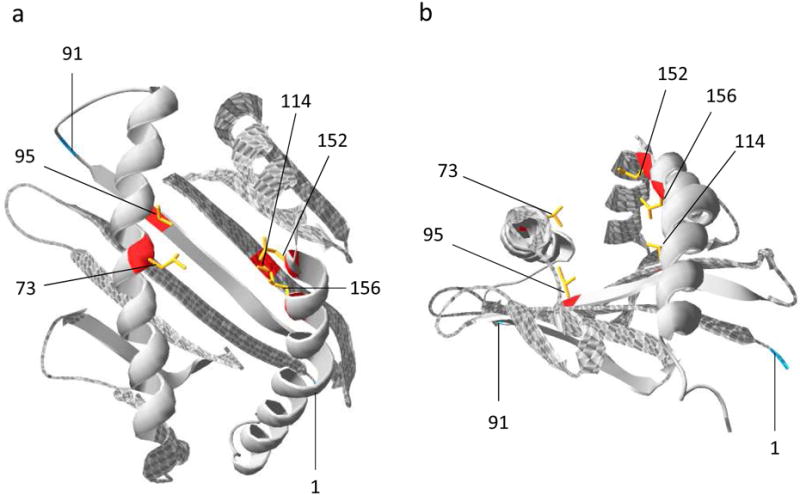
Schematic representation of DLA-88*03401 (α1 and α2 domains) showing positively selected amino acid sites. Nucleotide sequences of exons 2 and 3 from all DLA-88 alleles were aligned by codons and analyzed in two parallel runs using the software program MrBayes 3.1.2 (55). In each run, 1.0 × 106 cycles were performed. Posterior probabilities for each amino acid site were analyzed using the Tracer 1.5 program (56); codons were presumed to be under positive selection when posterior probability values exceeded 0.95. The burn-in value was set to the first 10% of cycles for each run. The proposed model is based on the high-resolution crystal structure of HLA-A*1101 (57), and was generated using SWISS-MODEL (58-60). Views show onto (A) and along (B) the peptide binding groove of the heavy chain (gray). Positively selected sites are indicated in red, with side chains shown in yellow. The first residues of the α1 domain (1) and α2 domain (91) are shown in teal.
Bottlenecks in population size – in this case, due to breed founding some 50 to 75 generations ago – result in the loss of genetic variation at polymorphic loci, which is seen as decreased allele numbers and heterozygosity (36). For example, marked restrictions in class I polymorphisms due to selective breeding are evident in miniature swine (30). To compare the allelic variation present in our sample groups, we used the Shannon entropy as an index of diversity. The Shannon entropy is a member of the family of valid diversity indices that equally rates species richness and clonal dominance (37). Entropy was estimated with correction for unseen species and inference of confidence intervals, as previously described by our group (38), which allows the statistical significance of entropy comparisons to be evaluated. Allelic diversity was not statistically different between Golden Retrievers (1.78, 95% confidence intervals [CI] 1.60 – 1.96) and Boxers (1.75, 95% CI 1.52 – 1.98); both populations were under-sampled for comprehensive discovery of all possible alleles (data not shown), as determined by the method of Egeland and Salas (39). Although the total number of alleles in the populations is unknown, this value can be estimated from the observed allele distributions. When this computation is performed using a non-parametric estimator (40), as implemented in the software package SPADE (41), the calculated number of DLA-88 alleles (and 95% CI) in the Golden Retriever and Boxer populations are 11.5 (10.1 – 25.1), and 10.3 (10.0 – 14.8), respectively. These values represent a level of allelic diversity at this classical MHC class I locus far less than that of humans, and suggest that our experiment was successful in recovering the major alleles from each population.
The finding of limited intrabreed diversity at the DLA-88 locus is not unexpected, as previous investigations of MHC class II allelic variation show analogous restrictiveness. For example, at the DLA-DRB1 locus, only 8 alleles were found in 31 Golden Retrievers, with 3 alleles (DLA-DRB1*01201, *01501 and *02001) being dominant (14). Similarly, 5 DLA-DRB1 alleles were observed in 41 Boxers; 71% shared the DLA-DRB1*00401 allele. The magnitude of DLA-88 restriction likely varies with breed. That Boxers have a single, dominant allele – DLA-88*03401 – while Golden Retrievers do not is presumably the result of a less severe bottleneck in the latter breed (42). Therefore, breeds with very few founders, such as the Basenji, Doberman, or Irish wolfhound, would be expected to carry few alleles. On the other hand, Beagles appear to have more class I variability. When the prevalence data from the 5 dogs in this study is combined with sequence-based typed Beagles in the literature (n = 6)(22) and (n = 2)(10), 10 different alleles (DLA-88*00101, *00401, *00402, *00501, *00601, *01602, *04501, *50101, *50201 and *50801) are observed in just 13 dogs. Interestingly, when diversity is calculated for the Beagle population, the confidence interval of the entropy estimation crosses that of the Golden Retriever and Boxer populations, although the interval is quite wide. This observation might be explained by increased species richness (more alleles) in Beagles in the setting of less equally distributed alleles, but additional work would be necessary to confirm this hypothesis. As a second measure of decreased intrabreed genetic variation at the DLA-88 locus, we also observed high levels of homozygosity in the sample dogs: 38.3% in Golden Retrievers; 35.9% in Boxers. While it is possible that our primer set failed to amplify some alleles, causing an overestimation of homozygosity, this seems less likely, as none of the 91 dogs in this study failed to return a DLA-88 sequence. Moreover, our data is consistent with the range of MHC homozygosity (33-40%) measured at the DLA-DRB1, -DQA1 and -DQB1 loci in several studies (14, 15).
As one might anticipate, interbreed variability for DLA-88 was high. Thus, of the 10 DLA-88 alleles identified in each breed, only 4 were shared, reflecting the different founders and genetic separation of these breeds. Moreover, the frequency distribution of alleles was dissimilar. For example, the two most commonly occurring variants in Golden Retrievers, DLA-88*05101 and *50801, had a prevalence of only 9.4% and 2.4%, respectively, in Boxers, while DLA-88*03401 was not observed in any Golden Retriever, despite being found in 82% of Boxers. Additionally, neither breed possessed two of the alleles that were present in our small sample of Beagles (DLA-88*00402 and *50201). These data suggest that the discovery of novel DLA-88 alleles could be expedited by genotyping dogs from different breeds, particularly by obtaining samples across genetic clusters (43), as has been done for canine class II (14).
The small sets of DLA-88 polymorphisms found in comparatively large cohorts of dogs allowed us to estimate the potential total number of alleles in these breed populations, which, as described above, appears unlikely to exceed 25 in Golden Retrievers and 15 in Boxers. An important caveat to this conclusion is that many (but not all) of the animals evaluated in this study came from a relatively restricted geographic location in the southeastern United States. In humans, it is well established that for polymorphic genes, such as HLA class I and II, the prevalence of alleles can vary widely by region (18). Hence, by sampling from a limited area, we may have underestimated allelic diversity. Comparisons of variability at class II loci between European and North American dogs of the same breeds do reveal similar allele prevalence and dominant species (15, 16), but a few non-shared alleles are also observed. Thus, genotyping Golden Retrievers and Boxers from other regions ultimately will be useful to corroborate our findings. Nor could we strictly verify that all dogs were unrelated. The majority of samples, however, were obtained from the NCSU-VTH, which is a large (>20,000 accessions per year) veterinary referral center for North Carolina, Virginia, South Carolina, and Tennessee, and therefore, most dogs were very unlikely to be related. For Boxer samples obtained from a regional breed association (n = 9), for whom pedigrees were available, none of the dogs had a common sire or dam in >4 generations. Conversely, while all dogs in this study appeared to be purebred by visual inspection (performed by one of the authors, PRH), we did not verify lineage in all individuals, and undocumented outbreeding could have led to an overestimation of class I diversity.
The restricted intrabreed diversity at the DLA-88 locus found in this investigation suggests that it may be practical to study antigen-restricted CD8+ T-cell responses by screening dogs drawn from these easily recognizable subsets for highly prevalent allele usage. There are also well-documented associations of breeds with predispositions to particular diseases, including those modulated or mediated by the immune system – infectious diseases, cancer, and autoimmunity – and ultimately, it may be possible to correlate specific DLA-88 variants with susceptibility or resistance, as has been accomplished with canine class II alleles (44-50). Lastly, peripheral hematopoietic stem cell transplantation is becoming an increasingly utilized treatment for canine lymphoma (51), and the limited diversity within breeds will undoubtedly facilitate haplotype matching to advance the use of allogeneic donors for this therapy.
Supplementary Material
Conservation of amino acids in the α1 and α2 domains between HLA-A2 (top sequence; GenBank accession AAB16923.1) and seven novel DLA-88 alleles in the β-2 microglobulin (b2m) and peptide binding (pb) sites, and at two conserved cysteine (cc) residues. Light gray residues indicate identities, while those in dark gray indicate differences from the human reference sequence.
Acknowledgments
We thank Jeff Thorne, Jeff Yoder, Adam Birkenheuer, Savannah Gabriel, Barb Hegarty, Victoria Catto, Don Creech, Poem Turner, Julie Fisher, Kim Williams, the NCSU-Veterinary Teaching Hospital Clinical Pathology Laboratory, and the Tar Heel Boxer Club of Greater Raleigh, Inc. for their assistance. This work was supported by a National Institutes of Health grant K08 DK082264 to PRH, and is dedicated to the memory of Chester Mahoney.
Abbreviations
- ARS
Antigen recognition site
- DLA
Dog leukocyte antigen
- HLA
Human leukocyte antigen
- HVR
Hypervariable region
- MHC
Major histocompatibility complex
- PBMC
Peripheral blood mononuclear cells
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest.
References
- 1.Robinson J, Waller MJ, Parham P, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–4. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham P, Adams EJ, Arnett KL. The origins of HLA-A,B,C polymorphism. Immunol Rev. 1995;143:141–80. doi: 10.1111/j.1600-065x.1995.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 3.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 4.Hill AV, Allsopp CE, Kwiatkowski D, et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 5.Siddle HV, Kreiss A, Eldridge MD, et al. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Natl Acad Sci U S A. 2007;104:16221–6. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett RC, DeRose SA, Wagner JL, Storb R. Molecular analysis of six dog leukocyte antigen class I sequences including three complete genes, two truncated genes and one full-length processed gene. Tissue Antigens. 1997;49:484–95. doi: 10.1111/j.1399-0039.1997.tb02783.x. [DOI] [PubMed] [Google Scholar]
- 7.Burnett RC, Geraghty DE. Structure and expression of a divergent canine class I gene. J Immunol. 1995;155:4278–85. [Google Scholar]
- 8.Graumann MB, DeRose SA, Ostrander EA, Storb R. Polymorphism analysis of four canine MHC class I genes. Tissue Antigens. 1998;51:374–81. doi: 10.1111/j.1399-0039.1998.tb02976.x. [DOI] [PubMed] [Google Scholar]
- 9.Wagner JL, Creer SA, Storb R. Dog class I gene DLA-88 histocompatibility typing by PCR-SSCP and sequencing. Tissue Antigens. 2000;55:564–7. doi: 10.1034/j.1399-0039.2000.550607.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardt C, Ferencik S, Tak R, Hoogerbrugge PM, Wagner V, Grosse-Wilde H. Sequence-based typing reveals a novel DLA-88 allele, DLA-88*04501, in a beagle family. Tissue Antigens. 2006;67:163–5. doi: 10.1111/j.1399-0039.2006.00497.x. [DOI] [PubMed] [Google Scholar]
- 11.Venkataraman GM, Stroup P, Graves SS, Storb R. An improved method for dog leukocyte antigen 88 typing and two new major histocompatibility complex class I alleles, DLA-88*01101 and DLA-88*01201. Tissue Antigens. 2007;70:53–7. doi: 10.1111/j.1399-0039.2007.00839.x. [DOI] [PubMed] [Google Scholar]
- 12.Vila C, Seddon J, Ellegren H. Genes of domestic mammals augmented by backcrossing with wild ancestors. Trends Genet. 2005;21:214–8. doi: 10.1016/j.tig.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Pang JF, Kluetsch C, Zou XJ, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–64. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy LJ, Barnes A, Short A, et al. Canine DLA diversity: 1. New alleles and haplotypes. Tissue Antigens. 2007;69(1):272–88. doi: 10.1111/j.1399-0039.2006.00779.x. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy LJ, Barnes A, Happ GM, et al. Extensive interbreed, but minimal intrabreed, variation of DLA class II alleles and haplotypes in dogs. Tissue Antigens. 2002;59:194–204. doi: 10.1034/j.1399-0039.2002.590303.x. [DOI] [PubMed] [Google Scholar]
- 16.Angles JM, Kennedy LJ, Pedersen NC. Frequency and distribution of alleles of canine MHC-II DLA-DQB1, DLA-DQA1 and DLA-DRB1 in 25 representative American Kennel Club breeds. Tissue Antigens. 2005;66:173–84. doi: 10.1111/j.1399-0039.2005.00461.x. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy LJ, Carter SD, Barnes A, et al. Interbreed variation of DLA-DRB1, DQA1 alleles and haplotypes in the dog. Vet Immunol Immunopathol. 1999;69:101–11. doi: 10.1016/s0165-2427(99)00046-x. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–9. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis MW. A plasmid Editor (ApE) 2011 [Google Scholar]
- 20.Ellis SA, Bontrop RE, Antczak DF, et al. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–8. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 21.Bird RC, Deinnocentes P, Lenz S, Thacker EE, Curiel DT, Smith BF. An allogeneic hybrid-cell fusion vaccine against canine mammary cancer. Vet Immunol Immunopathol. 2008;123:289–304. doi: 10.1016/j.vetimm.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Wagner JL, Sarmiento UM, Storb R. Cellular, serological, and molecular polymorphism of the class I and class II loci of the canine Major Histocompatibility Complex. Tissue Antigens. 2002;59:205–10. doi: 10.1034/j.1399-0039.2002.590304.x. [DOI] [PubMed] [Google Scholar]
- 23.Wellman ML, Krakowka S, Jacobs RM, Kociba GJ. A macrophage-monocyte cell line from a dog with malignant histiocytosis. In Vitro Cell Dev Biol. 1988;24:223–9. doi: 10.1007/BF02623551. [DOI] [PubMed] [Google Scholar]
- 24.Maloney S, Smith A, Furst DE, et al. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104:41–7. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass EJ, Oliver RA, Russell GC. Duplicated DQ haplotypes increase the complexity of restriction element usage in cattle. J Immunol. 2000;165:134–8. doi: 10.4049/jimmunol.165.1.134. [DOI] [PubMed] [Google Scholar]
- 26.Villegas-Castagnasso EE, Diaz S, Giovambattista G, Dulout FN, Peral-Garcia P. Analysis of ELA-DQB exon 2 polymorphism in Argentine Creole horses by PCR-RFLP and PCR-SSCP. J Vet Med A Physiol Pathol Clin Med. 2003;50:280–5. doi: 10.1046/j.1439-0442.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- 27.Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 28.Warburton RJ, Matsui M, Rowland-Jones SL, et al. Mutation of the alpha 2 domain disulfide bridge of the class I molecule HLA-A*0201. Effect on maturation and peptide presentation Hum Immunol. 1994;39:261–71. doi: 10.1016/0198-8859(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 29.Gu X, Nei M. Locus specificity of polymorphic alleles and evolution by a birth-and-death process in mammalian MHC genes. Mol Biol Evol. 1999;16:147–56. doi: 10.1093/oxfordjournals.molbev.a026097. [DOI] [PubMed] [Google Scholar]
- 30.Ando A, Kawata H, Shigenari A, et al. Genetic polymorphism of the swine major histocompatibility complex ( SLA) class I genes, SLA-1, -2 and -3. Immunogenetics. 2003;55:583–93. doi: 10.1007/s00251-003-0619-0. [DOI] [PubMed] [Google Scholar]
- 31.Tallmadge RL, Campbell JA, Miller DC, Antczak DF. Analysis of MHC class I genes across horse MHC haplotypes. Immunogenetics. 2010;62:159–72. doi: 10.1007/s00251-009-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foss V. Golden Retrievers Today. New York: Prentice Hall/Macmillan Co.; 1994. pp. 7–11. [Google Scholar]
- 33.Walker JH. The Boxer Handbook. Hauppauge, NY: Barron's; 2000. pp. 1–3. [Google Scholar]
- 34.Babiuk S, Horseman B, Zhang C, et al. BoLA class I allele diversity and polymorphism in a herd of cattle. Immunogenetics. 2007;59:167–76. doi: 10.1007/s00251-006-0173-7. [DOI] [PubMed] [Google Scholar]
- 35.Pokorny I, Sharma R, Goyal SP, Mishra S, Tiedemann R. MHC class I and MHC class II DRB gene variability in wild and captive Bengal tigers (Panthera tigris tigris) Immunogenetics. 2010;62:667–79. doi: 10.1007/s00251-010-0475-7. [DOI] [PubMed] [Google Scholar]
- 36.Allendorf FW. Genetic Drift and the Loss of Alleles Versus Heterozygosity. Zoo Biol. 1986;5:181–90. [Google Scholar]
- 37.Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–39. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- 38.Vincent BG, Young EF, Buntzman AS, et al. Toxin-coupled MHC class I tetramers can specifically ablate autoreactive CD8+ T cells and delay diabetes in nonobese diabetic mice. J Immunol. 2010;184:4196–204. doi: 10.4049/jimmunol.0903931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egeland T, Salas A. Estimating haplotype frequency and coverage of databases. PLoS One. 2008;3:e3988. doi: 10.1371/journal.pone.0003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao A. Species estimation and applications. In: Balakrishnan N, Read CB, Vidakovic B, editors. Encyclopedia of Statistical Sciences. Second. New York: Wiley; 2005. pp. 7907–16. [Google Scholar]
- 41.Chao A, Shen TJ. Program SPADE (Species Prediction And Diversity Estimation) 2010 Program and User's Guide published at http://chao.stat.nthu.edu.tw.
- 42.Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 43.Parker HG, Kim LV, Sutter NB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–4. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 44.Wilbe M, Sundberg K, Hansen IR, et al. Increased genetic risk or protection for canine autoimmune lymphocytic thyroiditis in Giant Schnauzers depends on DLA class II genotype. Tissue Antigens. 2010;75:712–9. doi: 10.1111/j.1399-0039.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 45.Wilbe M, Ziener ML, Aronsson A, et al. DLA class II alleles are associated with risk for canine symmetrical lupoid onychodystrophy [corrected](SLO) PLoS One. 2010;5:e12332. doi: 10.1371/journal.pone.0012332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy LJ, Barnes A, Ollier WE, Day MJ. Association of a common dog leucocyte antigen class II haplotype with canine primary immune-mediated haemolytic anaemia. Tissue Antigens. 2006;68:502–8. doi: 10.1111/j.1399-0039.2006.00715.x. [DOI] [PubMed] [Google Scholar]
- 47.Kennedy LJ, Davison LJ, Barnes A, et al. Identification of susceptibility and protective major histocompatibility complex haplotypes in canine diabetes mellitus. Tissue Antigens. 2006;68:467–76. doi: 10.1111/j.1399-0039.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy LJ, Quarmby S, Happ GM, et al. Association of canine hypothyroidism with a common major histocompatibility complex DLA class II allele. Tissue Antigens. 2006;68:82–6. doi: 10.1111/j.1399-0039.2006.00614.x. [DOI] [PubMed] [Google Scholar]
- 49.Quinnell RJ, Kennedy LJ, Barnes A, et al. Susceptibility to visceral leishmaniasis in the domestic dog is associated with MHC class II polymorphism. Immunogenetics. 2003;55:23–8. doi: 10.1007/s00251-003-0545-1. [DOI] [PubMed] [Google Scholar]
- 50.Angles JM, Famula TR, Pedersen NC. Uveodermatologic (VKH-like) syndrome in American Akita dogs is associated with an increased frequency of DQA1*00201. Tissue Antigens. 2005;66:656–65. doi: 10.1111/j.1399-0039.2005.00508.x. [DOI] [PubMed] [Google Scholar]
- 51.Willcox JL, Pruitt AF, Suter Autologous peripheral blood hematopoietic cell transplantation in dogs with B-cell lymphoma. J Vet Int Med. 2012 doi: 10.1111/j.1939-1676.2012.00980.x. In press. [DOI] [PubMed] [Google Scholar]
- 52.Drummond AJ, Ashton B, Buxton S, et al. Geneious. v.5.04. 2010. [Google Scholar]
- 53.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 54.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 55.Huelsenbeck JP, Dyer KA. Bayesian estimation of positively selected sites. J Mol Evol. 2004;58:661–72. doi: 10.1007/s00239-004-2588-9. [DOI] [PubMed] [Google Scholar]
- 56.Rambaut A, Drummond AJ. Tracer v1.5. 2007 Available from: http://beast.bio.ed.ac.uk/
- 57.Blicher T, Kastrup JS, Buus S, Gajhede M. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr D Biol Crystallogr. 2005;61:1031–40. doi: 10.1107/S0907444905013090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 59.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conservation of amino acids in the α1 and α2 domains between HLA-A2 (top sequence; GenBank accession AAB16923.1) and seven novel DLA-88 alleles in the β-2 microglobulin (b2m) and peptide binding (pb) sites, and at two conserved cysteine (cc) residues. Light gray residues indicate identities, while those in dark gray indicate differences from the human reference sequence.


