To the editor
Urban et al.1 recently challenged the dendritic nucleation model of lamellipodia protrusion based on their inability to detect branched actin filaments in lamellipodia by electron tomography. We have carefully analysed the primary data that was provided as Supplementary Information by Urban et al., and report that there are numerous branches that have been overlooked by the authors.
Lamellipodia are thin veil-like protrusions of the cell edge that have important roles in cell migration. Their advance is driven by the polymerization of actin filaments. The exact mechanisms of lamellipodia protrusion are still debated, owing in part to controversy regarding the structural organization of the actin filaments in lamellipodia. Initial structural data obtained by Small et al.2 using negative-staining electron microscopy suggested that lamellipodia contained a network of long diagonally oriented actin filaments. Based on this structure, the authors proposed a treadmilling model of lamellipodia protrusion stating that actin filaments continuously elongate at their distal barbed (plus) ends, thus pushing the membrane, and continuously depolymerize from the proximal pointed (minus) ends, thus maintaining actin turnover.
A large body of structural, biochemical, kinetic and functional data accumulated over the subsequent three decades has led to a revised model of lamellipodia protrusion, termed the dendritic nucleation model3. A key additional point of this model is that the actin filaments in lamellipodia are constantly nucleated by the Arp2/3 complex as branches on pre-existing filaments. However, despite the compelling evidence, Small and colleagues have questioned the dendritic nucleation model in a recent paper1. In their study, they challenge key evidence supporting the dendritic nucleation model; namely, visualization of branched actin filaments in lamellipodia by platinum-replica electron microscopy4,5. Urban et al. argue1 that the branched configuration of the actin filaments in these samples are an artefact of critical-point drying, a part of the sample preparation. To prove their hypothesis that the actin filaments in lamellipodia are long and unbranched, they analysed the structure of lamellipodia by cryo-electron microscopy, and did not detect branched actin filaments in lamellipodia1.
Because the data from platinum-replica electron microscopy that demonstrate branched actin filaments in lamellipodia have been reported primarily by our group, and because we have received multiple requests from the scientific community to comment on the results of Urban et al., we would like to share our opinion with a broader audience through this commentary. Several points, including comparison of potential artefacts produced by the two electron microscopy techniques and evaluation of evidence for and against the dendritic nucleation model, have already been addressed6,7. However, one critical point has not been discussed; namely, the fundamental consistency between our results and those of Urban et al. in spite of the different interpretations.
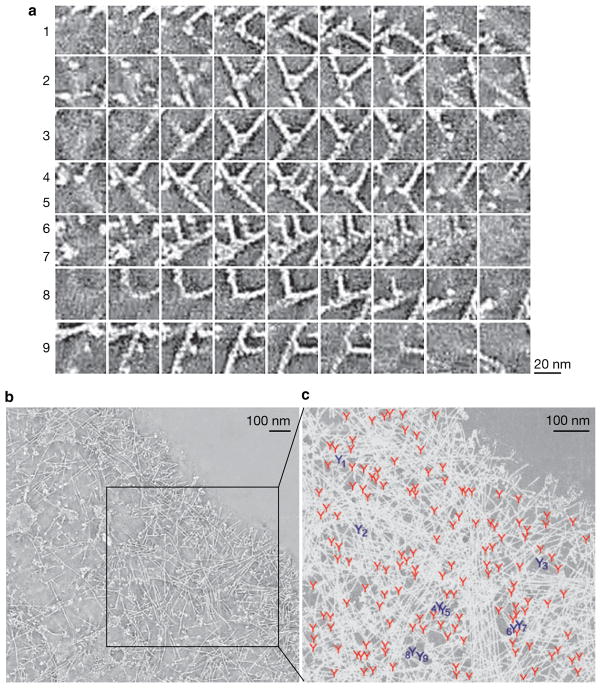
We have carefully analysed the primary data provided as Supplementary Information by Urban et al. Based on this analysis, we argue that a subset of their results provides strong supporting evidence for the dendritic nucleation model by showing branched actin filaments in lamellipodia. We have found that Supplementary Movie S6, showing an electron tomogram of the lamellipodium in a 3T3 cell, is of particularly good quality to illustrate this point. Several examples of branched actin filaments from this movie are shown in Fig. 1a as a montage of z planes. In these examples, one of two actin filaments never re-emerges on the other side of the second filament in adjacent movie frames, despite the fact that both filaments are in the same z plane in at least one frame of the series. These features demonstrate that the end of the former filament makes a contact with the side of the latter filament, but does not cross it, which is the definition of a branch. In the zoomed region of the movie that shows a cell area of ~0.53 μm2, we have found a total of 147 branches (Fig. 1b), which is in contrast to the authors’ statement that “branches at the sides of actin filaments were extremely rare”. Branched actin filaments can be also found in other Supplementary movies, but fewer branches can be clearly detected in these movies owing to their insufficient quality and selection of regions corresponding to filopodia or filopodial precursors, which contain long unbranched filaments.
Figure 1.
Branched actin filaments in the lamellipodium of 3T3 fibroblast. The figure is adapted from Supplementary Movie S6 by Urban et al.1 that shows a scan through an electron tomogram generated from a two-axis tilt series of electron microscopy images. (a) Examples of individual branches (numbered at left). For each branch, a montage of adjacent z planes in the tomogram is shown. Some frames contain two branches (numbered 4,5 and 6, 7). Note a distinct blob at each branch. (b) Single plane of the full-field tomogram with box indicating the zoomed region shown in c, which we used for identification of branches. (c) Positions of all detected actin branches marked by Y symbols are shown on the background of the maximum projection image of the region of the tomogram shown in the boxed region in b. Numbered Y symbols in dark blue indicate positions of branches shown in a with corresponding numbers. Scale bars are 100 nm, as deduced from Supplementary Fig. S4 by Urban et al. showing a fragment of this cell.
Remarkably, all branches identified in our analysis invariably contained a blob at the branch point that probably corresponds to the Arp2/3 complex. Furthermore, the angle between branched filaments was 70 ± 7 degrees (n = 57), consistent with the conventional angle of ~70° produced by the Arp2/3 complex in vitro8. The actin filament branches found in data from the Urban et al. paper are virtually indistinguishable by appearance from those obtained by Hanein using cryo-electron microscopy of actin branches reconstituted in vitro from the Arp2/3 complex and actin, and could even be fitted into the three-dimensional branch model developed in these studies9.
In conclusion, existence of branched actin filaments in lamellipodia is supported by two different electron microscopy techniques that probably counterbalance their potential artefacts. We are not sure why Urban et al. failed to detect branches, but this is definitely an issue of seeing or not seeing, rather than a problem with the techniques. Thus, in our opinion there should be no controversy regarding the structural organization of actin filaments in lamellipodia, and the dendritic nucleation model can serve as a conceptual framework for subsequent studies in the field.
Acknowledgments
We thank R. Dominguez, M. Ostap and M. Lampson for their helpful comments on this Correspondence. This work was supported by NIH grants GM70898 to T.S.
Footnotes
AUTHOR CONTRIBUTIONS
C.Y. and T.S. analysed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 2.Small JV. J Cell Biol. 1981;91:695–705. doi: 10.1083/jcb.91.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 4.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svitkina TM, Borisy GG. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgs HN. Trends Cell Biol. 2011;21:2–4. doi: 10.1016/j.tcb.2010.09.010. author reply 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Insall RH. Trends Cell Biol. 2011;21:2. doi: 10.1016/j.tcb.2010.11.002. author reply 4–5. [DOI] [PubMed] [Google Scholar]
- 8.Mullins RD, Heuser JA, Pollard TD. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanein D. Methods Enzymol. 2010;483:203–214. doi: 10.1016/S0076-6879(10)83010-1. [DOI] [PubMed] [Google Scholar]