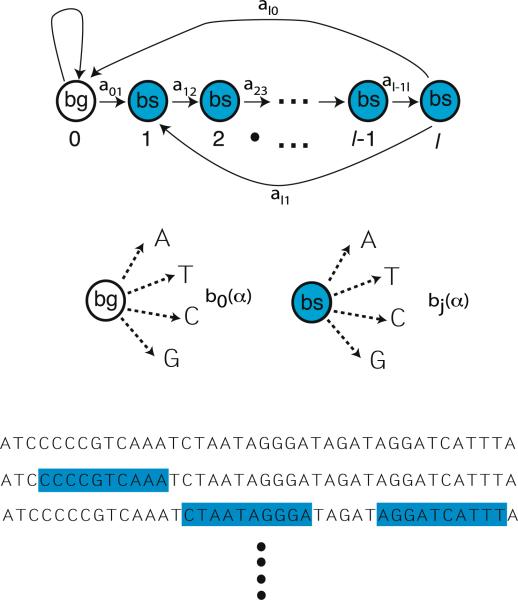
Fig. 1.
Hidden Markov Model for binding sites of size l. (Top) There are l + 1 hidden states, with state 0 background DNA and state j corresponding to position j = 1 . . . l in a binding site. The HMM is described by a Markov process with transition probabilities give by aij . (Middle) Each state j in an HMM is characterized by an observation symbol probability bj (k), for the probability of seeing symbol k = A, T , C, G in a state j. (Bottom) A given sequence of DNA is composed of binding sites and background DNA