Figure 1.
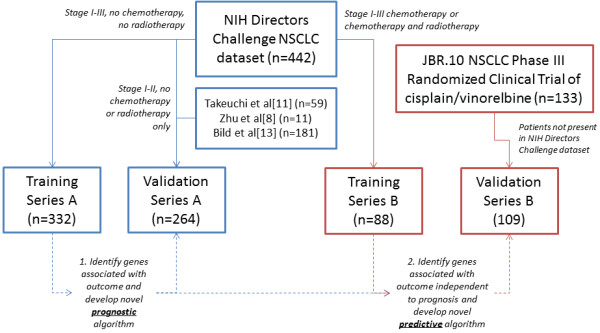
Schematic diagram of datasets used to form training and validation series used in this study. Data from treatment-naïve adenocarcinoma patients enrolled in the NIH Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma were first used to develop a prognostic signature able to predict DSS, independent to clinical factors such as age and clinical stage [10]. This signature was validated on the independent adenocarcinoma series listed and then used to identify a new set of genes from ACT-treated patients that were associated with outcome, independent to prognosis. The second algorithm (ACT-response) was validated on data from Zhu et al. [8].
