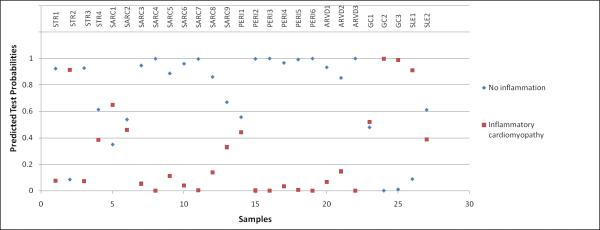
Figure 3. Prediction Analysis of Microarrays (PAM) applying the developed molecular signature for inflammatory cardiomyopathy in patients with secondary cardiomyopathy (n=27).
The transcriptomic biomarker performed with 100% sensitivity and 95% specificity in identifying inflammation in patients with stress induced cardiomyopathy (STR, n=4), sarcoidosis (SARC, n=9), peripartum cardiomyopathy (PERI, n=6), arrhythmogenic right ventricular dysplasia (ARVD, n=3), giant cell myocarditis (GC, n=3) and systemic lupus erythematosus (SLE, n=2). One patient with STR (sample #109) and another one with SLE (sample #76) were identified as inflammatory cardiomyopathy. Indeed, when results from immunohistochemistry were revised, those 2 samples contained significant lymphocytic infiltrates. One sample from the group with sarcoidosis (sample #113) was misclassified as inflammatory cardiomyopathy, while the report from histopathology revealed no signs of inflammation. All samples from patients with giant cell myocarditis were correctly identified.