FIGURE 1.
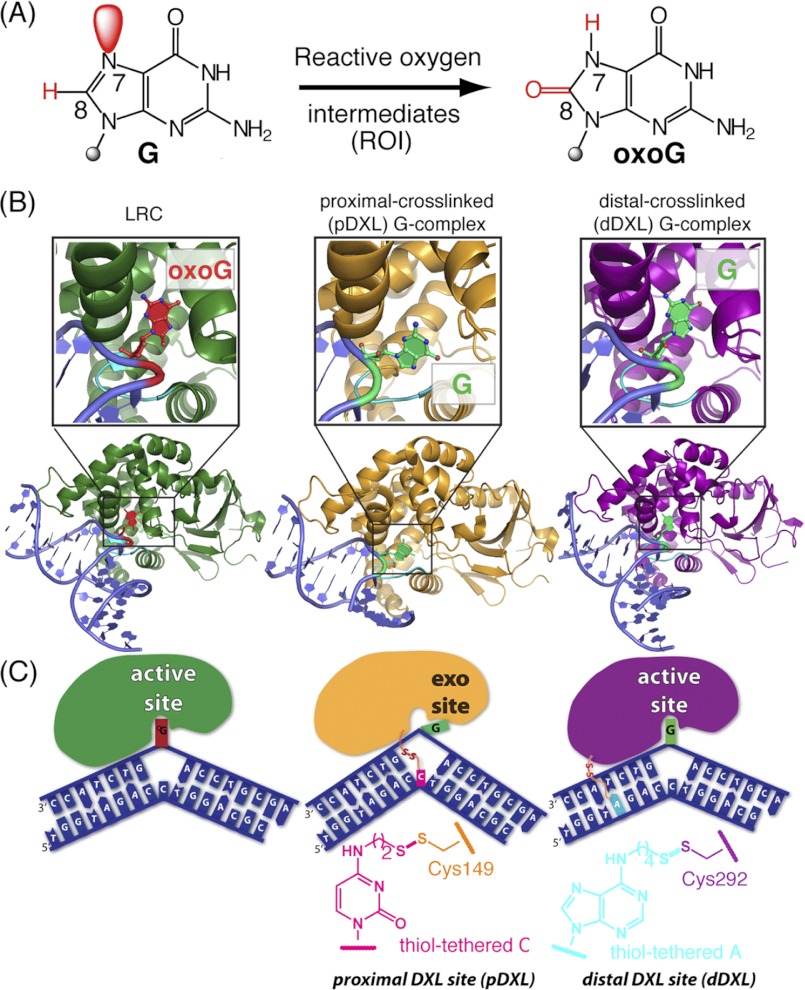
Overview of hOGG1-DNA complexes and disulfide cross-linking strategies. A, chemical structures of guanine and 8-oxoguanine, with the constitutional differences at the 7 and 8 positions highlighted in red. B, schematic representations of: left, a lesion recognition complex (LRC, 1EBM (10)) obtained using a recognition-competent but catalytically inactive mutant of hOGG1 bound to DNA, with oxoG (red) fully inserted into the active site lesion-recognition pocket; middle, the proximally disulfide cross-linked (pDXL) G-complex (orange, 1YQR (34)), with the target G (green) in the exo-site; and right, distally dDXL G-complex (3IH7, present work) of hOGG1 bound to normal DNA, with the target G (green) in the active site. The DNA interrogation loop containing the conserved NNN motif is colored cyan. C, schematic representations of the structures in B. In the pDXL and dDXL structures, the protein is wild-type, except for Cys residues engineered for purposes of cross-linking (N149C and S292C, respectively). Chemical structures of the tethered nucleobases employed in cross-linking are shown.