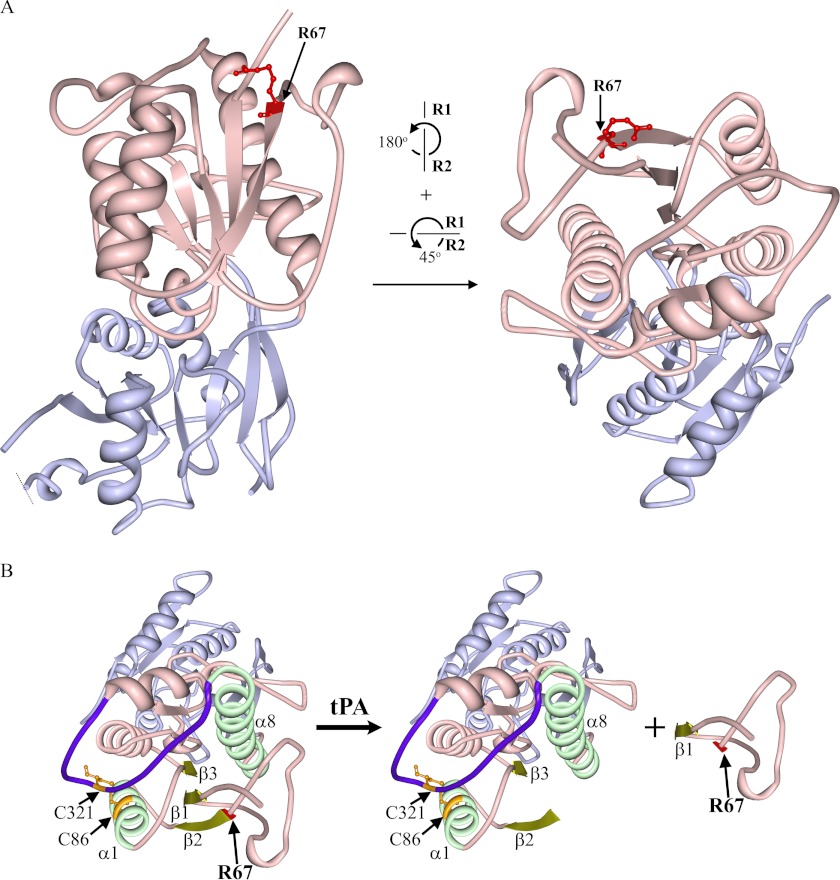
FIGURE 5.
Crystal structure of apoNR2BATD. A, the strand representation of the crystal structure of apoNR2BATD (PDB code 3JPW) (47), which consists of two domains, R1 (light pink) and R2 (light purple) was created with Protein Workshop (60). Arg67 (R67; residue highlighted in red with balls and sticks) is situated at the tip of R1 (light pink) and thus is proposed to be exposed to the surrounding aqueous milieu. Left, shown is the front view of the NR2BATD crystal structure. Right, shown is a view of the crystal structure when the protein molecule is rotated as denoted. B, several structures that may be critical for the tertiary structure of the ATD upon the removal of Gln29–Arg67 are highlighted. The tPA cleavage site Arg67 (R67) is highlighted in red; Gly36–Val42 fragment (β1 sheet), Val68–Met73 (β2 sheet), and Val97–Asp101 (β3 sheet) are highlighted in dull yellow; Pro78–Asp91 (α1 helix) and Leu289–His311 (α8 helix) are highlighted in light green. The hypervariable loop (HVL) is highlighted in dark purple, and the two cysteine residues, Cys86 in α1 and Cys321 in HVL (highlighted in orange with balls and sticks), form a disulfide bond that helps to stabilize the ATD structure. The peptide upstream of Val68 can dissociate from the receptor after tPA treatment.