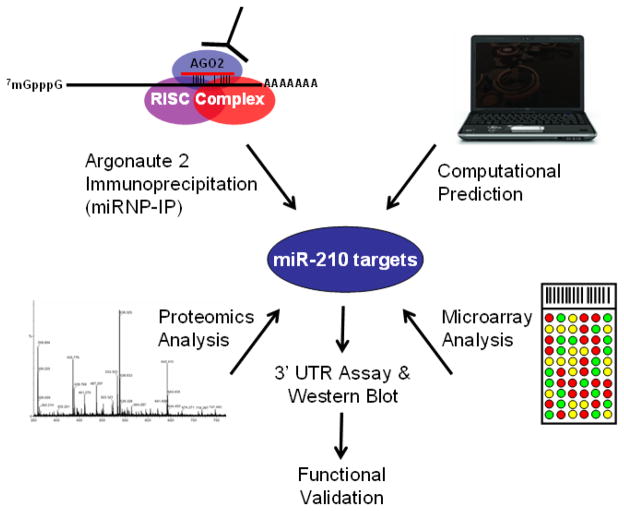
Figure 2.
Strategies for identifying miR-210 targets. Schematic view of four major strategies utilized to identify miRNA targets. By immunoprecipitating a major component of the RISC complex, Argonaute 2 protein (AGO2), miRNP-IP pulls down the mRNA targets associated with miRNA within RISC. The mRNA targets are identified by next-generation sequencing or microarray. Because miRNA regulates target gene expression through the inhibition of RNA translation or degradation of target mRNA, two strategies can be used to identify miRNA targets, proteomics analysis or mRNA microarray profiling. When miRNA targets identified through experimental approaches are combined with computational prediction, which is mainly based on the homology of the short miRNA “seed region” to the 3′ UTR sequence of target gene, a list of candidate miRNA target genes are generated. 3′ UTR sequences of the putative targets are cloned into a reporter construct and a luciferase reporter assay is then performed to measure inhibition of protein production. Western blotting is also performed to verify decreased protein levels. The verified miRNA target gene is then subject to appropriate functional assays for validation.