Abstract
We introduce two concepts: the Query Web as a layer of interconnected queries over the document web and the semantic web, and a Query Web Integrator and Manager (QI) that enables the Query Web to evolve. QI permits users to write, save and reuse queries over any web accessible source, including other queries saved in other installations of QI. The saved queries may be in any language (e.g. SPARQL, XQuery); the only condition for interconnection is that the queries return their results in some form of XML. This condition allows queries to chain off each other, and to be written in whatever language is appropriate for the task. We illustrate the potential use of QI for several biomedical use cases, including ontology view generation using a combination of graph-based and logical approaches, value set generation for clinical data management, image annotation using terminology obtained from an ontology web service, ontology-driven brain imaging data integration, small-scale clinical data integration, and wider-scale clinical data integration. Such use cases illustrate the current range of applications of QI and lead us to speculate about the potential evolution from smaller groups of interconnected queries into a larger query network that layers over the document and semantic web. The resulting Query Web could greatly aid researchers and others who now have to manually navigate through multiple information sources in order to answer specific questions.
Keywords: Ontologies, data integration, semantic web, query web
1. Introduction
In recent years researchers spend as much or more time at their computers as they do in the wet lab. A significant component of this time is spent managing experimental or clinical research data with the aid of spreadsheets, local databases, or small-scale web-based databases [1], [2], [3], [4]. In addition, with the worldwide increase in web-accessible databases at levels ranging from chemicals to public health [5–25] researchers tend to spend significant time searching these databases for information that is available about, for example, a particular gene, protein or disease of interest. In fact more often than not researchers first consult the online databases before doing experiments, often in order to develop hypotheses that can be tested in the lab.
However, because most databases are designed independently, with different data models and (generally web-based) user interfaces, it has become an increasingly significant effort for researchers to access and combine data from different sources. In a 2002 paper [26] Stein likened the situation to a nation of city-states, with the concomitant problems of lack of standards and methods of access. The immediately obvious solution of a single worldwide repository and/or a universal agreed on set of standards is not compatible with the dynamic nature of biomedical research, and would therefore stifle the innovation that is the source of excitement and progress in the field. In his paper Stein advocated for the use of web services to provide computer-readable access to databases. In the intervening years such services have become increasingly available. However, accessibility is only part of the requirement for data integration. Other requirements include interoperability and an integration architecture.
Interoperability is increasingly being addressed by semantic web technologies, including the linked-data initiative [27], which in particular has led to the vision of a “global database” of linked data and knowledge. However, at the present time most data and knowledge are not yet linked to each other, and although often accessible via web services, are represented and queried in different ways. What is relatively common, however, is that most data can be expressed in some form of XML, which along with JSON [28], is a common method for exchanging data. Thus, if ways can be found to integrate different forms of XML and/or JSON data then at least part of the vision of the global database can be achieved while waiting for the linked data initiative to become more widespread.
Integration of such XML-based data requires an integration architecture in addition to accessibility and interoperability. Multiple such architectures have been developed over the years; ranging from heavyweight, which require significant effort to set up and maintain, with the gain of increased reliability and quality; to lightweight, which, like web pages, are easy to setup, but may not be easily reusable or shareable [29].
One heavyweight data integration approach described by Goble [29] is called “view integration”. Views are very popular in relational databases because they permit complex relational tables to be virtually joined into new tables that are more tailored to the interests of particular users. The virtual tables are not actually materialized in the database, but are generated by saved queries. The primary conceptual idea that makes views work is that they exhibit closure, in which the output of the view query is of the same format (a relational table) as the input, thereby permitting views to be treated as source data. SQL queries over the view tables look to the query writer like any other queries; however, at runtime these queries are composed with the view queries to generate queries over the underlying materialized tables. Underneath the hood considerable optimization is employed to make these queries run faster, including caching of the results of running the view queries.
The “view integration” approach extends this notion to the Internet, whereby relational tables are analogous to remote sources accessed via web services, and the “view” over these sources is a central mediated schema [30]. The mediated schema is associated with a mediator application that reformulates queries over the mediated schema to calls to the remote services, some of which may be ontologies (or the mediated schema may itself be an ontology).
As in relational databases such an approach has appeal because it allows the user to only understand the mediated schema, which may be highly customized to his or her own interests; the mediator does all the work of understanding the potentially large number of source schemas. However, in spite of these potential advantages, most systems have only been deployed in laboratory settings [31, 32], and there are only a few examples of mediated systems in widespread use [18, 33] because it is very expensive to maintain the mediated schema and to change it as the underlying source schemas change. Thus, like data warehouses, only well-funded groups can support this approach.
In this paper we describe a data integration architecture that attempts to retain the advantages of the view integration approach while remaining lightweight. Like relational views we take advantage of the closure property such that queries generate results that can themselves be queried, thus allowing queries to be treated as views. However, rather than relational tables, the closure is over XML (with potential extensions to JSON as query languages are developed for that representation [34]). We describe our Query Integrator (QI) application, which permits any user to create, save and reuse a query over any web accessible source(s) in whatever language is most appropriate for the source. The only requirements are that the sources are either in some form of XML or generate some sort of XML as output, and that the results of the saved query also generate some sort of XML. Because of this closure property the saved query can be executed via a (possibly parameterized) REST web service, thereby permitting it to be treated like any other query able web source, and permitting saved queries to be chained together. The ability to chain queries over other saved queries allows anyone to create an arbitrarily complex view over any number of data sources. Like views over relational data the view can present information to users in ways more tailored to their interests, and that hide unnecessary detail.
Once such queries have been saved they can become available via our REST service as yet another Internet web service that can be included in custom applications that include end-user graphical interfaces, or into a workflow system like Taverna [35], which allows further processing, analysis and visualization of the results from the query.
The approach we describe is not designed for end-users, who would most likely prefer to fill out forms in a custom application that accesses the saved queries under the hood. Rather, our system is primarily intended for experts in the query languages and sources, who would then make their saved queries available for use by other experts through modifications of saved queries, or by end-user applications that access the saved queries via the REST service.
In the remainder of this paper we describe the architecture of the Query Integrator (QI), illustrate its use for a detailed use case in neuroimaging, show current saved queries that address several additional use cases, and discuss research issues that still need to be resolved. We end by speculating how a Query Web could evolve by linking together multiple instances of the Query Web Integrator and Manager, and how such a query web could be accessed by applications that help end-users more easily integrate data within the rapidly evolving field of biomedicine.
2. System Design
2.1. System architecture
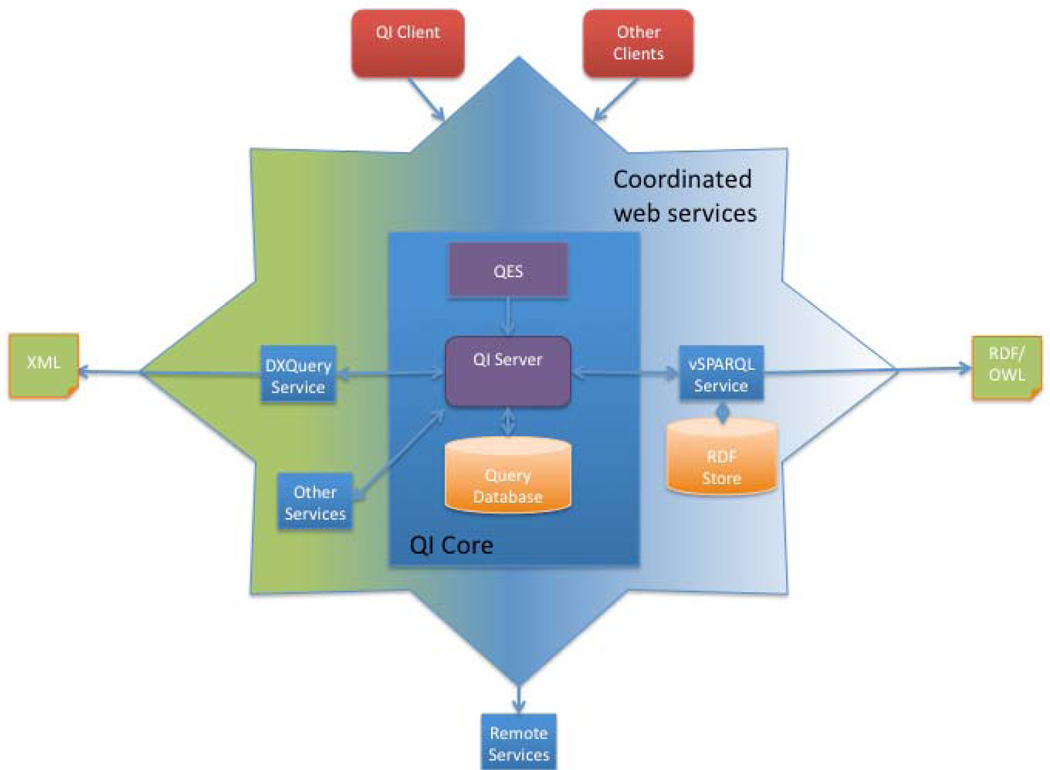
As shown in Figure 1 the Query Integrator and Manager (QI) consists of three basic components: QI Core, which includes a server (QI Server), Query Database and REST Query Execution Service (QES); a graphical user interface web client written in Adobe Flex (QI Client); and an expandable set of coordinated query web services (DXQuery Service, vSPARQL Service and Other Services in the figure).
Figure 1.
Query Web Integrator and Manager (QI) architecture. QI consists of a set of core components and a set of coordinated web services. Core components include the Query Database, QI Server, and Query Execution REST Service (QES). Current coordinated web services include query services for each supported language (e.g. the DXQuery Service over XML, the vSPARQL service over RDF/OWL, and other services as described in the text). QI can also access remote services and be accessed by its own as well as other clients either through the QES service or via Action Message Format (AMF) calls directly to the QI Server.
In general the user interacts with the QI Client to create, save and edit web queries in any of several supported languages; the resulting queries are stored in and retrieved from the query database by the query server; and execution of the queries from either the QI Client or the REST service causes the server to dispatch them to the appropriate query service and to gather the results for presentation to the user.
The remainder of this section describes these components in more detail.
2.2. Core
The core components of QI consist of the query database, server and query execution REST service.
2.2.1. Query database
QI stores queries and their associated metadata in a relational database, currently PostgreSQL [36]. The most important relations in the Query Database are the User, Query, and Graph tables. The User table contains information about registered users of the system. This information is used for authorization and discovery. Individual users of QI are authorized to view, edit, and execute any of their own queries. Additionally, users can view and execute the shared (public) queries of others. To aid in query discovery, user information can also be used as search criteria.
The Query table stores the actual queries as well as any associated metadata, such as title, description, language, owner, public/private, etc. Each query is also assigned a unique id. This id is an essential feature of the system and will be discussed further in section 2.2.3.
The Graph table stores information about data sources registered with the system. These sources may be internal to the QI system, which supports the upload of query results as new materialized sources. The sources may also be external, pointers (URLs) to useful sources and metadata about those sources. Information about registered sources is used primarily to inform users of data sources available for query.
2.2.2. QI Server
The QI Server communicates with the Flex client, query execution service, query database, and query services to facilitate query and query metadata storage, sharing, discovery and execution. It uses the Hibernate object relational mapping library to mediate the exchange of objects between the database and the server. Query execution is delegated to the appropriate query services (presently, but not limited to, SOAP web services) according to the query’s “language” metadata tag.
The QI server API, which is accessible to any application, supports these basic query operations. Both the Flex client (section 2.3) and the Query Execution Service (section 2.2.3) use this API. Outside clients may access the server as well, for example our VIQUEN graphical query generator [37].
2.2.3. Query execution service
The Query Execution Service (QES) is a REST web service, which executes a query based on its query ID. The QES interacts with the QI server, which in turn interacts with the appropriate query processing service to evaluate the identified query. In addition to basic query execution given a query ID as part of the URL, the service also supports parameterized queries through additional arguments to the REST URL. Parameterization schemes includes direct substring substitution, regular expression substitution, and variable substitution, in which numbered placeholders in a template query are replaced by sequential arguments from the URL. Template queries are illustrated extensively in section 3.
While the QES is relatively simple, it is a very important component of the overall QI system. Since the current query processing services can process a document retrieved from a URL, and since the QES REST services are accessible via URL, a query saved in the QI database can access another QI query, either in the local installation or in a remote installation. A call to the QES looks like a request for a static document, but in actuality it is resolving a query on-the-fly and returning the results as a document. By chaining or otherwise combining such queries (e.g. by referring to a QES URL from another query) we can build up more complex queries. These parts may be written in different query languages, choosing the language best suited for a particular sub-task. This ability to chain queries together potentially leads to the evolution of the Query Web as we discuss further in section 5.
2.3. User Interface
The QI client provides the necessary interfaces for performing a variety of query management tasks. The primary interface (Figure 2) allows users to enter queries and edit certain query metadata fields (title, description, and language). The possible values currently available in the “language” pull-down are those we describe in section 2.4: XQuery, DXQuery, SPARQL, vSPARQL, IML, moduleConfig and URL.
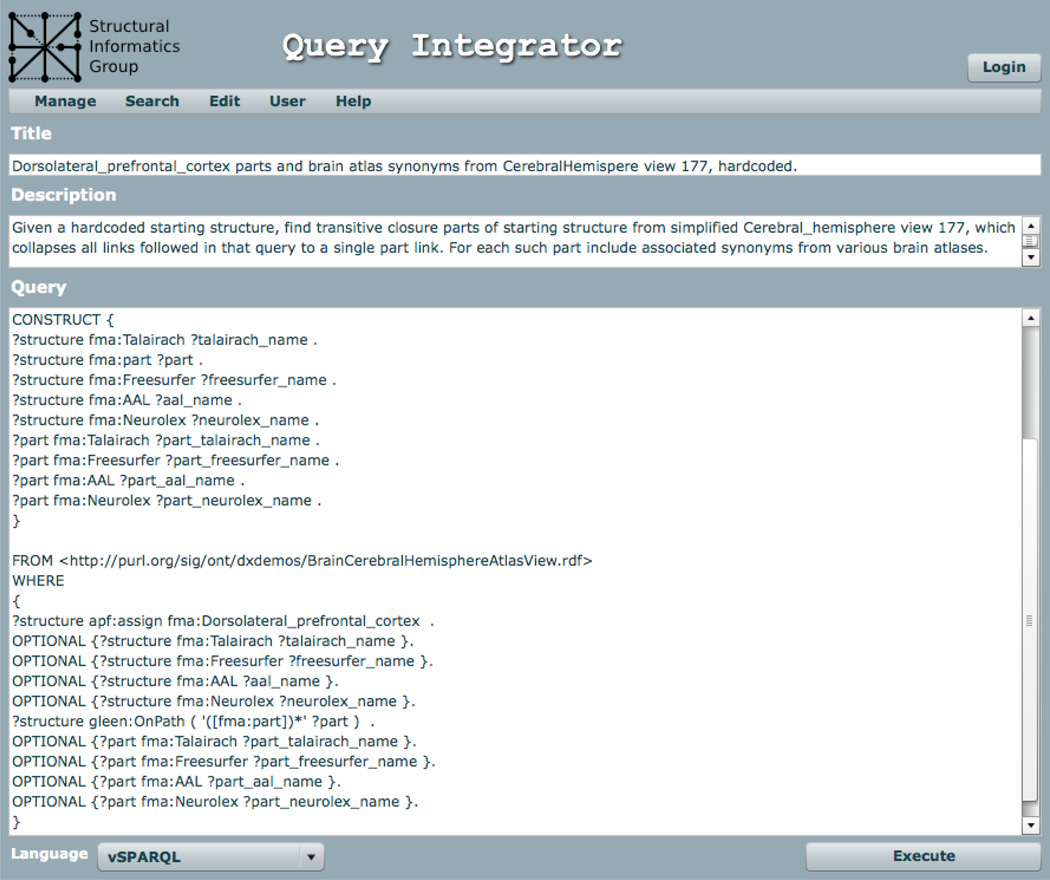
Figure 2.
The QI Client is written in Adobe Flex and runs in a standard web browser. Pull-down menus along the top allow the user to Manage, Search and Edit queries, to access User functions such as register, login and logout, and to receive Help. The primary information areas of a query retrieved from the Query Database are the Title, Description and the Query itself. Along the bottom of the user interface are a pull-down to select the Language the query is written in (or to display the language of a retrieved query), and a button to Execute the query and display the returned results. The particular query shown (query 178) is described in section 3.2. The only difference between this query and template query 179 as described in that section is in the second line following the WHERE keyword: in 179 “?structure apf:assign fma:Dorsolateral_frontal_cortex” is replaced by “?structure apf:assign <#0>”, where <#0> is a template variable to be filled in by the caller.
Queries can also be executed from this page. Additional interface components are available through the menus, including dialogs for searching for queries and registered graphs, saving queries and results (both to the local file system and to the QI server), and logging in existing or registering new users.
2.4. Query services
Presently the QI server communicates with 5 query processing web services, though it is extensible and can be configured to work with others. Query engines in the QI system are currently compartmentalized, for modularity, into distinct web services (presently SOAP services). Such a configuration has 2 main advantages: 1) a modular design makes extension easier as we add new query engines, and 2) QI can potentially communicate with query engines written in different programming languages (though the current services are all Java).
The current query processing services accessed by the QI are able to execute queries in the following languages: XQuery, DXQuery (distributed XQuery), SPARQL, vSPARQL (view SPARQL), IML (intermediate language), and two convenience “languages”. The first convenience language describes an ontology modularization task, whereas the second allows the developer to construct and save a complex URL, primarily to set parameters for REST service calls. The details of these query services are described in the following sections.
2.4.1. XQuery and Distributed XQuery
The distributed XQuery processor (DXQP), for accessing data in XML, is a service that processes standard XQueries as well as distributed XQueries (DXQueries) using extensions to XQuery that we developed [38]. The latter are like regular XQueries, but support calls to extension functions for interacting with other SOAP and REST services. The most common such services provide remote query processing at a document’s location, enabling query shipping, rather than document shipping (e.g. only the query and its results are exchanged on the web, not the entire data file). DXQuery is a component of our earlier DXBrain neuroscience data integration system [39] that we extracted for inclusion in QI.
2.4.2. SPARQL and vSPARQL
The vSPARQL query engine enables access to data in RDF and OWL by processing both standard SPARQL queries as well as queries written in our extended vSPARQL syntax [40]. vSPARQL extends SPARQL with both nested and recursive queries. Additionally it provides skolem functions enabling the dynamic creation of named RDF resources. The addition of these extra features makes it more suitable than standard SPARQL for specifying complex ontology views.
In addition to supporting direct RDF and OWL document access, this service supports query access to data stored in the persistent storage mechanism SDB provided by the Jena RDF framework [41]. SDB does not require an RDF or OWL document to be loaded or shipped in its entirety in order to process queries. In this way it is better suited to large files.
In our current vSPARQL deployment we have used SDB to provide efficient query access to several large ontologies or data sources, including our Foundational Model of Anatomy (FMA) [42], the National Cancer Institute Thesaurus (NCIt)[43], Reactome [44], and others. In addition to these hand chosen sources, we have persisted most of the RDF and OWL ontologies from the NCBO BioPortal (BP), a public repository for biomedical ontologies [45].
2.4.3. Other Services
The three services currently included in the “Other Services” box in Figure 1 are IML, and the two convenience services: Module Extraction and URL.
Our intermediate language, IML [46], is intended to be equivalent to vSPARQL in expressivity, but more intuitive for query composition. The basic expressions in IML were chosen to closely align with the sorts of operations that an ontology author/editor uses when manually extracting (and possibly modifying and/or extending) an ontology subset. The IML service does not directly evaluate incoming queries. Instead, it rewrites IML queries into equivalent vSPARQL queries and sends them to the vSPARQL service for evaluation. IML also lends itself more readily than vSPARQL to query optimization as we have demonstrated in [46], but have not yet incorporated in the IML service. It is well suited for use in a graphical query construction interface, as for example our VIQUEN application [37].
The first “convenience” query service in the current QI system is not really a query processor at all, at least not in the classic sense. Instead it is a web service providing logical module extraction from ontologies. An ontology module is a subset of a larger ontology for which the same conclusions hold (both asserted and inferred) with regard to a set of signature classes. We describe this service in more detail in section 4.1.
The second convenience “query service” allows the user to create and save a complex URL as an executable query. Since the URLs to many REST services may require complex arguments that often need to include escape characters, it can be very tedious to have to retype the URL each time the service is accessed. Thus, the “URL” language tag allows the user to save a URL in the Query Integrator, and then to retrieve and “execute” the URL just like any other query. We use this capability extensively in accessing our own REST services, as we describe in section 3.
2.5. Implementation
The above components are currently installed on two RedHat Linux server machines in the UW Structural Informatics Group (SIG): xiphoid, which runs an Intel 2.13 Ghz Xeon E5606 (8M cache) processor with 4G Ram; and axon, which runs an Intel 2.33 Ghz Core2 E6550 (4M cache) processor with 4G RAM. All components are currently written in Java and deployed on Tomcat [47], although the web services could be written in any language. The central QI query database is PostgreSQL, and currently contains over 208 queries, all stored as strings. The sizes of the separate service databases are described in section 4.
3. System Use
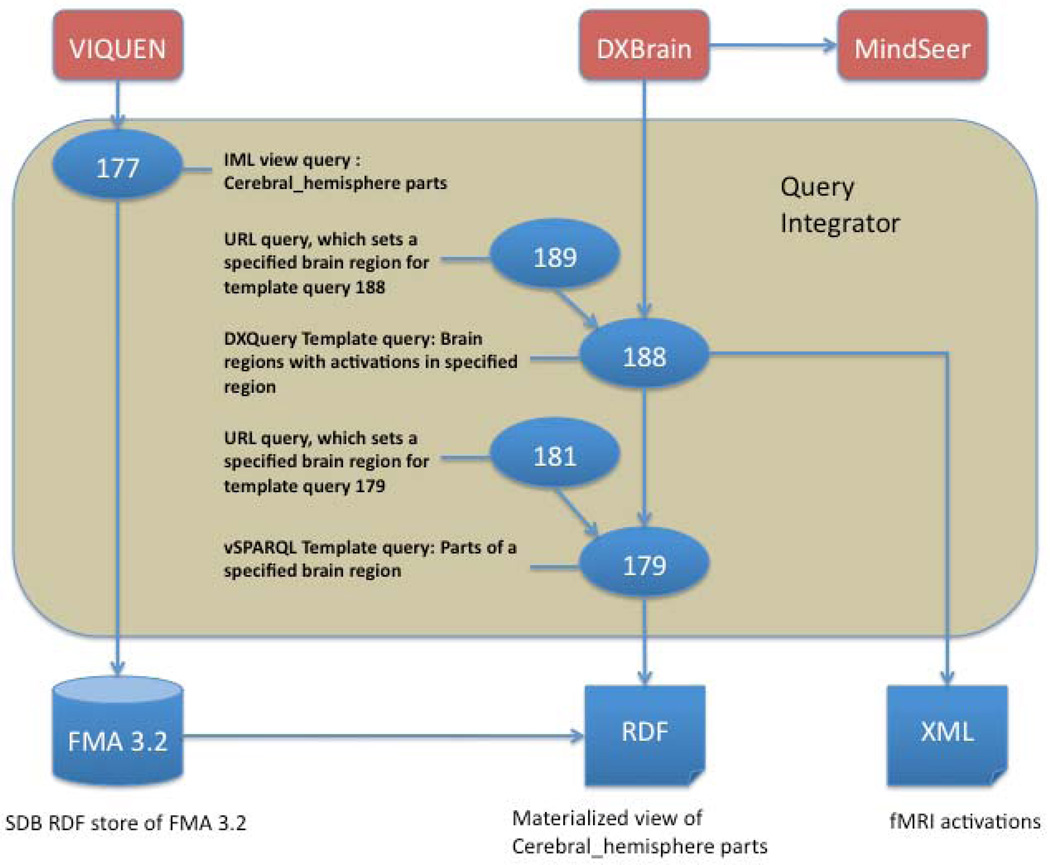
In this section we illustrate the basic QI operations in the context of a neuroimaging scenario, in which the goal is to combine a simplified view of a reference ontology with an XML data source in order to perform “intelligent” querying over a set of fMRI activation data. As shown in Figure 3 the data flow for this scenario takes advantage of the query chaining capability of QI in order combine queries in three languages (IML, vSPARQL and DXQuery). Specifically, we first use our visual query generator VIQUEN to generate IML query 177, which creates and saves a materialized neuroanatomical view of the Foundational Model of Anatomy (FMA) [42]. We create vSPARQL template query 179 over this view that, when accessed via the QES, returns the parts of any brain structure given as a parameter to the template query. We then use the results from this query to restrict the region of interest in a separate XML fMRI dataset to only those activations that are annotated with some part of the given brain structure (query 188). This query is in turn accessed by an outside application (DXBrain [39]) to allow display of activations on a 3-D brain by our Mind Seer application [48].
Figure 3.
Dataflow for the scenario described in section 3. Bottom row are data sources: FMA 3.2 saved in an SDB RDF data store, an RDF materialized view of FMA 3.2 saved as a file, and an XML file of fMRI activations. The top row is a set of external applications that interact with the Query Integrator. Numbered ovals refer to QI saved queries that either access the data sources directly or access other saved queries.
All queries in this and the next section, as well as our current installation, are available through the project web page [49]. Further references to these queries are by query ID.
3.1. Creation of a materialized Cerebral_hemisphere view
Smaller views of large ontologies like the FMA are intended to make the ontologies more useful for applications and to permit modular development. For OWL-DL a standard approach is module extraction, which we provide through our module extraction service as described in section 4.1.
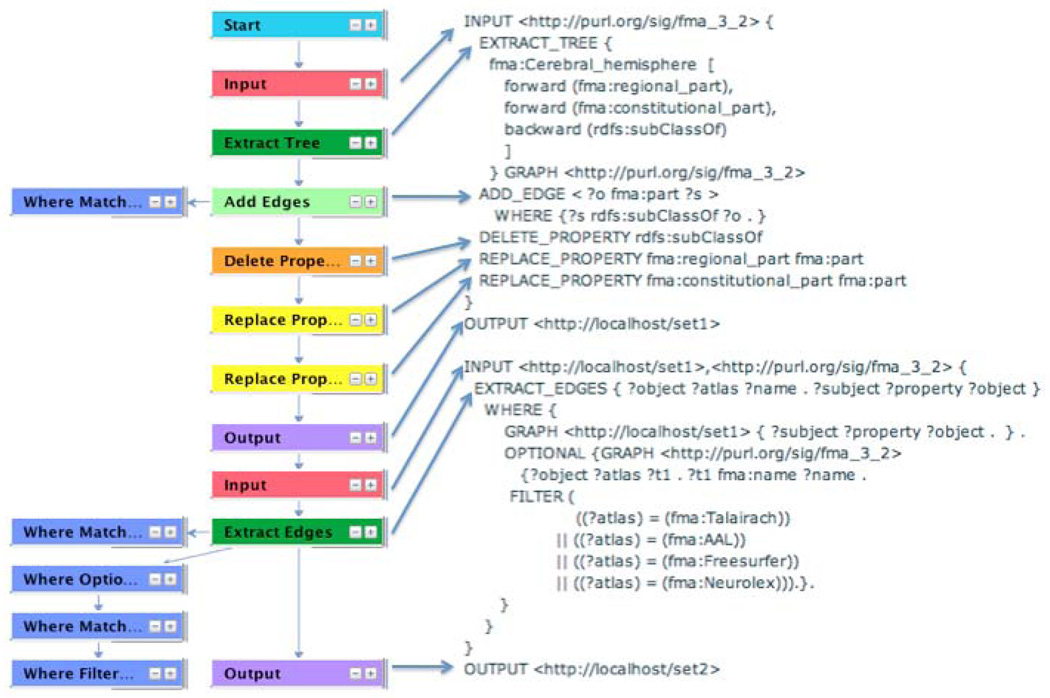
However, module extraction generates ontologies that are proper subsets of the parent, whereas in this case we want a view of the FMA that is not a strict subset but is easier for the user to understand and to query. Therefore, we used the VIQUEN visual query engine [37] to graphically generate an IML query, which in turn generates a simplified view of the FMA for use in relating different neuroanatomy parcellation schemes (Figure 4). The IML query generated by VIQUEN was copied into QI as query 177, with a few small edits since VIQUEN is not yet integrated with the QI. This query extracts a tree rooted at fma:Cerebral_hemisphere by following all rdfs:subClassOf, fma:regional_part and fma:constitutional_part links, and then converts all these links to fma:part. For each such part it also extracts synonyms from several standard brain parcellations schemes that are used in various brain atlases.
Figure 4.
Use of VIQUEN to generate IML view query 177. Screenshot of VIQUEN graphical layout on left, generated IML query on the right. Each box in VIQUEN represents an operation on an RDF graph, where the output graph of one operation is the input graph to the next, in a manner similar to a dataflow language. Once the graphical dataflow is designed (left) it is compiled to IML (right) where the correspondence between the boxes and the generated IML is indicated by arrows. Execution of the IML query by QI compiles the query to vSPARQL, runs the vSPARQL query over the data store (in this case fma_3_2 as defined by the top level INPUT operation), and returns the RDF result.
Execution of this query takes about a minute, so we saved the results of the query as a materialized view, shown as the RDF box at the bottom of Figure 3. Note that this view is no longer a “proper” ontology since all three of the primary relations have been collapsed into a single “part” relation. However, this simplified view is much easier for users to understand than the full FMA, and it suffices for the particular data integration application we demonstrate in the next section.
3.2. Generic brain parts
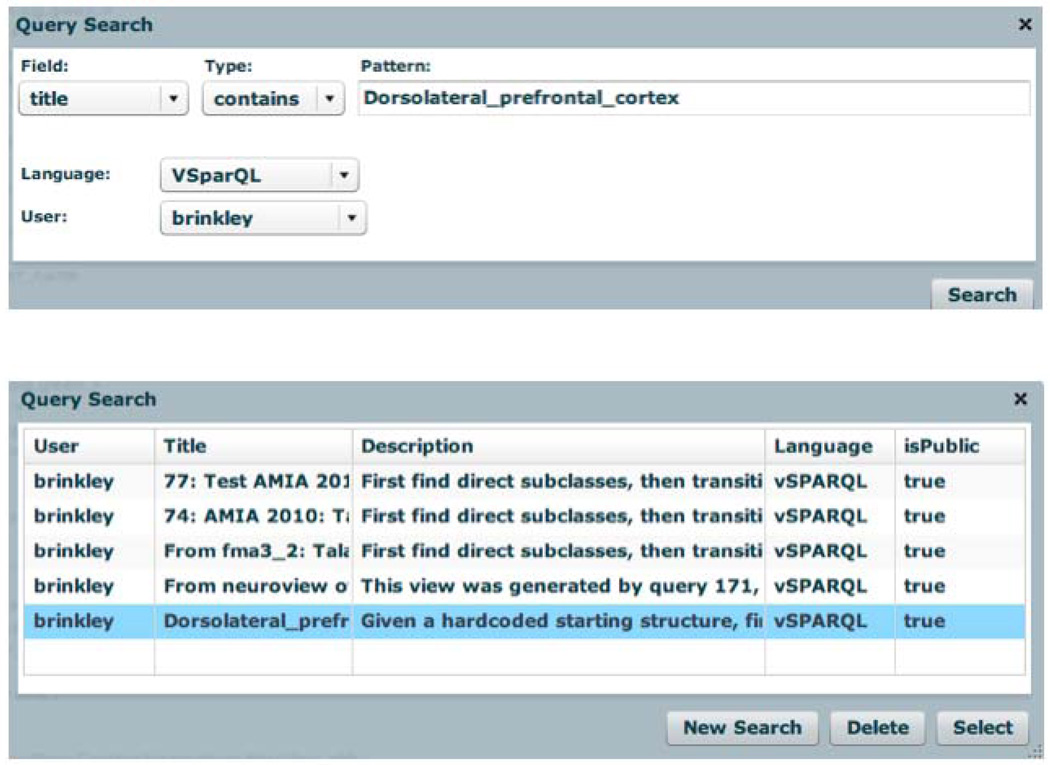
We next wrote a vSPARQL query over this view to determine the parts of a brain structure such as the Dorsolateral_prefrontal_cortex. (DLPFC). Initially we wrote a query to directly find the parts of the DLPFC, but we wished to modify that query to be more general. To do that we first needed to find the saved query since there are now over 208 saved queries in the query database. Figure 5 (top) shows the Query Search panel that appears when the user clicks on the “Query Search” menu item in the main interface (Figure 2). In this case we are looking for a vSPARQL query owned by user Brinkley, which has the string “Dorsolateral_prefrontal_cortex” somewhere in the title. Figure 5 (bottom) shows that as of this writing 5 saved queries satisfy these constraints. Clicking on the last query listed brings up the actual query, which is query 178, as shown in Figure 2. Note that this query is much simpler than a query over the full FMA would be, since we need only follow a single fma:part link type.
Figure 5.
Query search. Top: Search panel that pops up when the user clicks the Search menu in Figure 2. Bottom: Saved queries that satisfy the search constraints. The highlighted query is selected.
We then edited this query to find the parts of an arbitrary brain structure, and saved it as a new query 179. The only difference between this query and query 178 is that in query 179 the hardcoded structure name fma:Dorsolateral_prefrontal_cortex is replaced by a variable <#0>, making this a template query that must be executed by the QES. Query 181 is a URL saved query that calls this template query with parameter fma:Temporal_lobe (Figure 6). That is, it finds the parts and associated brain atlas synonyms for the temporal_lobe.
Figure 6.
Saved query 181. This is a query with language tag “URL”, which simply sets the single argument for template query 179. Executing this query calls the QES REST service, which returns the parts of the temporal lobe, based on the simplified view of the FMA created in view query 177. The query can either be executed from the QI client (Figure 2) by clicking the “Execute” button, or by simply pasting the URL into a standard web browser.
3.3. Intelligent queries over brain imaging data
We now show how the result of the above FMA template query 179 can be used by a query over data to perform “intelligent” queries, in this case returning all fMRI activation sites that are annotated by Talairach labels that are located in any part of a given brain structure. The Talairach labels are terms used in the Talairach neuroanatomic atlas [50], which is one of the standard parcellations schemes we have included as synonyms of FMA terms in the Cerebral_hemisphere view created by query 177 (section 3.1). The query we develop integrates two types of information sources: a knowledge source (ontology) representing the brain parts view of the FMA, which is stored in RDF and accessed via vSPARQL template query 179 over the Cerebral_hemisphere view, and an fMRI data source expressed in XML, which is accessed via XQuery template query 188.
The fMRI dataset is part of a multi-institutional study undertaken by the BIRN consortium, in which each of ten institutions performed the same fMRI tasks and protocols on healthy and schizophrenic subjects [51]. Acquired and processed data at each site were stored locally but made accessible via a federated grid system that makes all the data appear to be located in a single database. (This “heavyweight” system was very difficult to maintain, with the result that the BIRN project has recently taken a new direction, with a new lead institution [18]).
In previous reports we showed the use of the FMA and our earlier DXBrain application to query these data [52, 53]. As part of these efforts the fMRI image volumes from a simple calibration task were downloaded from the BIRN federated database, registered to the Montréal Neurological Institute (MNI) standard space [54], and analyzed to determine centers of brain activation as X-Y-Z MNI coordinates. These coordinates were shipped to the Talairach Demon [55], which automatically labeled each site with a Talairach anatomical label. The resultant labeled sites were then exported as an XML file, a portion of which is shown in Figure 7. Each site indicates whether it was from the healthy (h) or schizophrenic (s) Diagnostic_group, the number of contiguous voxels in the activation site, the normalized (MNI) coordinates (right_coord, sup_coord, ant_coord) of the voxel with maximal activation within the contiguous region, and the Talairach label of that maximal voxel as added by the Talairach demon.
Figure 7.
A single fMRI activation site, out of a total of 1741 in the XML file, with 842 from the schizophrenic group and 899 from the healthy group. Query 192 computes these statistics.
Saved XQuery 192 (not shown) simply adds a color to each site (red for healthy, yellow for schizophrenic) so they can be visualized.
3.4. Filtering by part of the brain
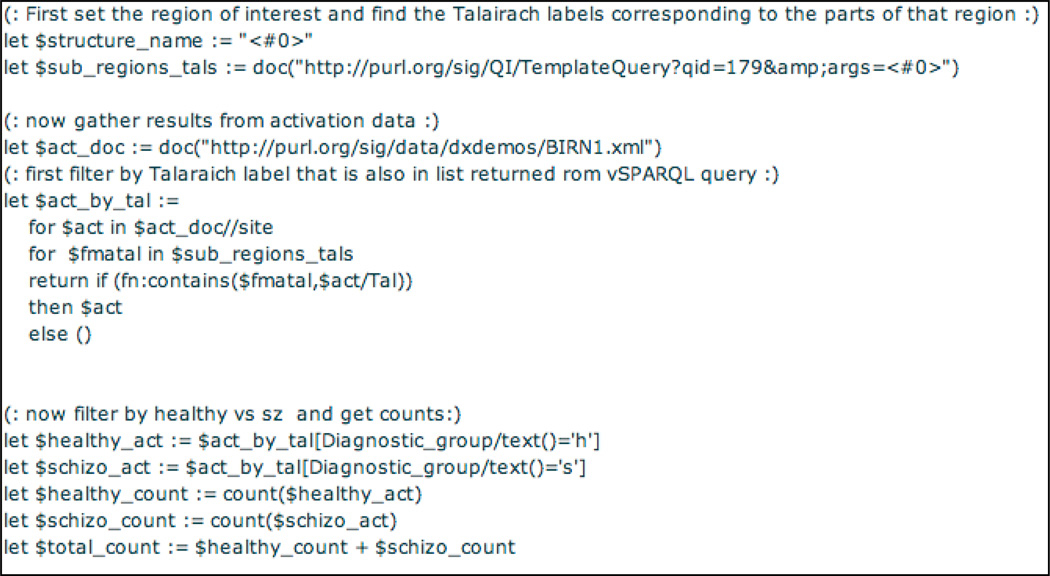
Template XQuery 188 (the first part of which is shown in Figure 8) references template query 179 (the ontology query defined in the previous section), via the XQuery doc function (line 3). The argument value passed in for template parameter <#0> is in turn passed along as a value for parameter <#0> in query 179, thus demonstrating the ability of one template query to pass a parameter to another template query.
Figure 8.
Snippet of XQUERY template query 188 over a SPARQL template query. The third line calls FMA SPARQL template query 179 to find the parts and Talairach labels of a brain structure given as a parameter. The activation data, a snippet of which is shown in Figure 7, is read in, and then filtered such that only those sites that have Talairach labels in the selected brain region are retained. Those sites that remain are then color coded according to diagnostic group in the rest of the query (not shown).
The results of this query are assigned to variable $sub_regions_tals in Figure 8. Each site in the original XML file is then checked to see if its Talairach label matches one of the labels returned by the brain parts query 179. If so the site is retained, if not it is rejected. The result of executing this query is that only sites in the specified region are retained. The rest of the query (not shown) simply adds a color to each site. Saved URL query 189 (not shown) sets the parameter for template query 188 to be fma:Dorsolateral_prefrontal_cortex. Execution of this query returns only those activation sites in the DLPFC (55 out of a total of 1741), where each site is in the same format as the site in Figure 7, with the addition of a color tag to indicate whether the site was from a healthy (25 sites) or schizophrenic (30 sites).
3.5. Visualizing via an external application
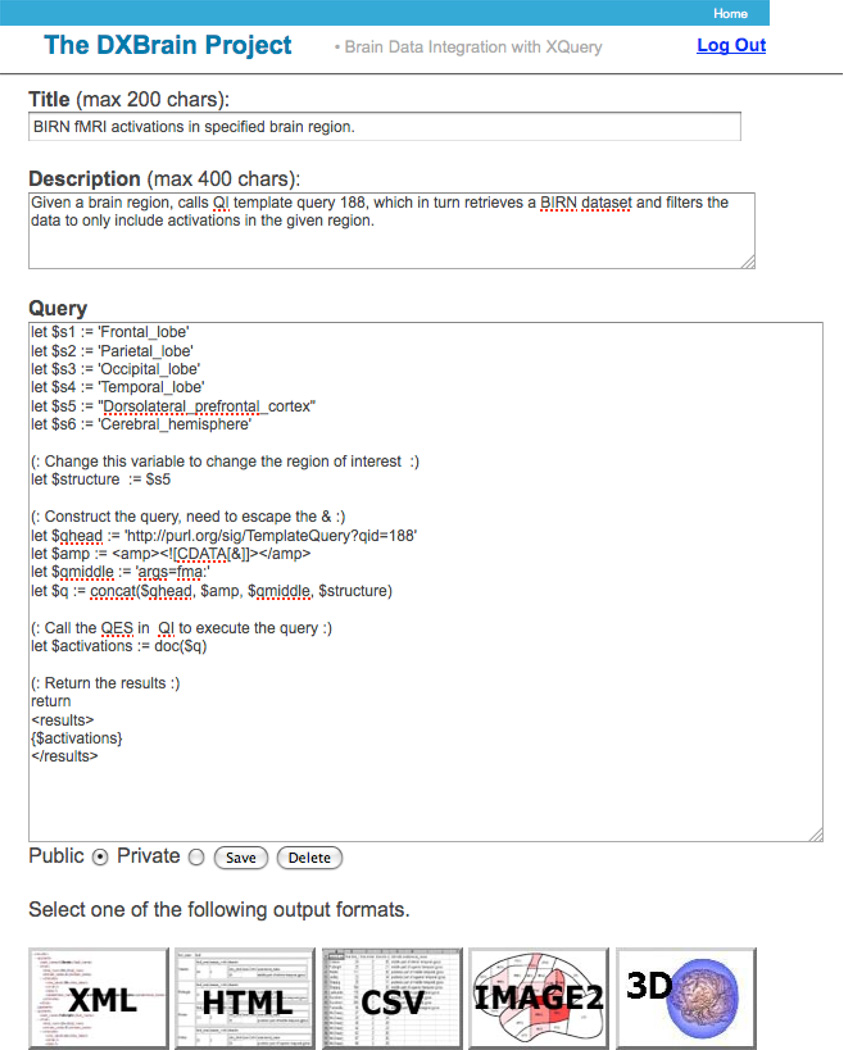
Template query 188 can be executed by the QES REST service from any client, not just the QI client. In particular Figure 9 shows our earlier DXBrain application accessing saved template query 188, with parameter fma:Dorsolateral_prefrontal_cortex. This $structure parameter can easily be changed to one of the other listed structures by simply changing the assignment statement. It would also be relatively straightforward to generate a web form that allows a user to select one of these parameters by clicking on a radio button, as we showed in our report on DXBrain [39].
Figure 9.
Our earlier DXBrain application was the inspiration for QI, but it is limited to storing only XQUERYs and doesn’t include a QES. However, since any XQUERY can access a REST service through the “doc” command, it does allow access to a saved query from QI. In this case it is accessing QI query 188, with parameter determined by the “$structure” variable, which is assigned to one of the enumerated variables at the top of the query. The output of the query can be visualized in multiple ways, as indicated by the buttons at the bottom of the screenshot.
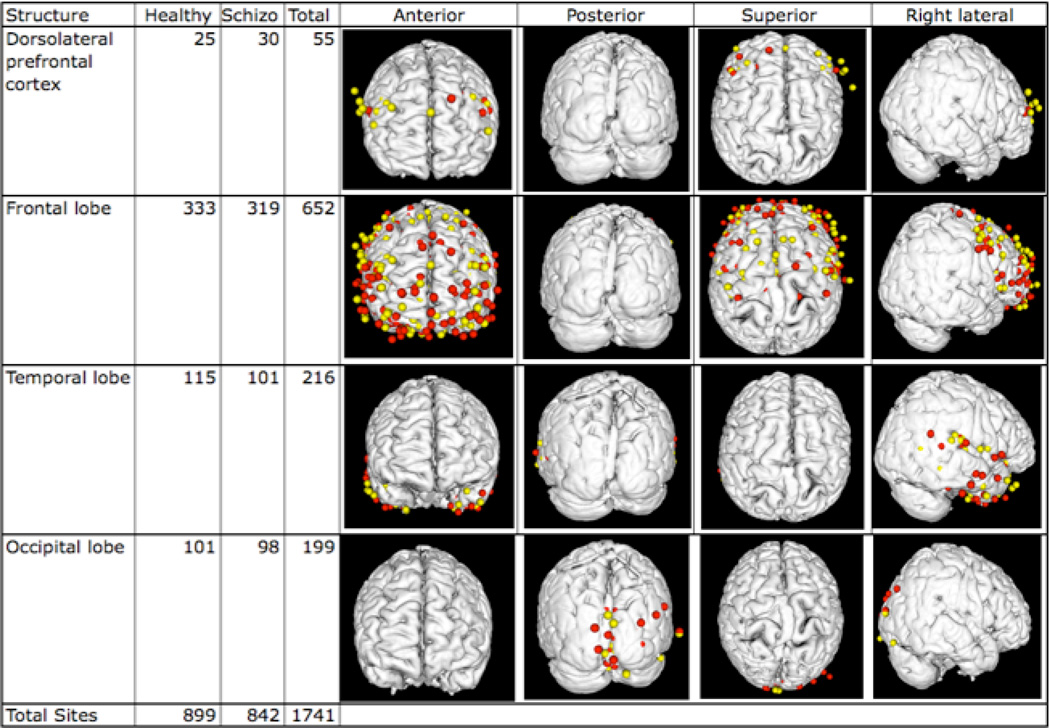
Although we plan to include plugins in QI that allow results to be viewed in different ways, QI currently only displays results as XML. DXBrain, on the other hand, allows the output results to be visualized in multiple ways by clicking one of the buttons on the bottom row. Results can be visualized as XML, HTML, CSV, a 2-D schematic brain image, and as a 3-D visualization. In the latter case the results are automatically piped to our Mind Seer application [48], which displays the activation sites on a standard brain (the Colin Atlas [56]). By simply changing the anatomical structure in the DXBrain query different activations in different regions of the brain can be visualized. For example, Figure 10 shows activations and counts in several of the structures listed in Figure 9. In each case, using the color parameters set in query 188, Mind Seer displays schizophrenic data in yellow and healthy data in red. For this particular functional imaging task there were no differences between schizophrenics and healthy controls, but other tasks in the original BIRN study did show differences [51]. By simply changing the XML source to be the results of one or more of those tasks wherein a difference was detected (which could itself be generated automatically through an analysis pipeline like NiPype [57] wrapped as a REST service that returns XML) and by changing the region of interest it should be possible to very rapidly explore different datasets and different regions of the brain.
Figure 10.
Some results from the DXBrain query shown in Figure 9. Each row is the result of changing the assignment of the $structure variable to one of the static structure names listed at the top of the query. The “Structure” column in each row is the structure name. The “Healthy” column lists the number of sites from subjects in the healthy group that were located within any part of the structure, as defined by the FMA and the Talairach labels in the data. Similarly, the “Schizo” column lists sites from subjects in the schizophrenic group, and the “Total” column lists the total number of sites in the region. The images are snapshots of the 3-D visualization tool Mind Seer from different viewpoints, where the colored dots are the results returned by the query. Red dots are from the healthy group, yellow are from the schizophrenic group. The bottom row shows the total number of sites in the dataset, which is not the sum of the sites in each row since only a few structures are shown.
4. Current Status and Additional Example Use Cases
The queries shown in Figure 3 represent 5 out of a total of 208 queries (a number that continues to increase) that are currently stored in our local instance of the QI database. The number of users listed in the user table is 17, all of whom are informaticists, and most of them are associated with our local group as of this writing. We have not yet tried to solicit outside users, although anyone can register and create queries.
The current saved queries span a range of use cases as illustrated on the supplemental web page [49], including bioinformatics database search (queries 108, 109 and 205); query-based extraction of ontology views similar to the Cerebral_hemisphere view (queries 50, 194 and 195); logic-based ontology module extraction (queries 130 and 203); value set extraction for use in clinical Electronic Data Capture (queries 130, 132, 135, 197, 198 and 203); image annotation using an ontology-based terminology service (query 95); remote execution of procedures for quality control of neuroimaging data (queries 97 and 102); local integration of clinical research data (queries 93 and 202); wide-scale integration of cancer research data (query 157); wide-scale integration of clinical research metadata via a single instance of QI (queries 163 and 164); and (simulated) wide-scale integration of clinical research metadata via multiple instances of QI (queries 164, 208, 209 and 210).
The supplemental web page shows the execution time for these queries, which range from 0.37 to 175 seconds, with a mean of 15 and a median of 4 seconds. This time is dependent on the number and size of the underlying source databases, the complexity of the queries, the number of queries in a query chain, the manner of storage of the source data (e.g. an XML file or an RDF triple store in these examples), the specific query engine(s) accessed by a query service, any optimizations that have been implemented in the query engine, and (for large queries such as the neuroanatomy view) the order of operations specified in the query. For these examples the size of the input XML files ranged from 2.7 to 2692K, whereas the number of triples in the SDB triple store database ranged from 333 to 3292K. With the exception of the neuroanatomy view (175 seconds) all these execution times were deemed to be in the acceptable range, but as the size of the data stores becomes much larger we will need to investigate methods for scaling up. One possibility is local caching of query results, and there are many others in the database literature.
In the following sections we highlight some of these use cases.
4.1. Logic-based ontology module extraction (queries 130 and 203)
The IML example presented in section 3.1, as well as other view queries shown on the supplemental web page (queries 50, 194 and 195) are examples of the graph-based approach to ontology view generation, which treats the source as a collection of nodes and edges (the underlying RDF graph) and extraction is based on patterns of connectivity within this structure [58]. This approach also supports the generation of output artifacts that are not "proper" ontologies and/or are not strictly subsets of the input ontology, as is the case for the view created in section 3.1. This capability may be needed in specific applications depending on the use case. An alternative method for view generation is the logical approach, which treats the source as a collection of logical axioms (description logic represented in OWL), and extraction is based on what can be inferred from a given axiom collection [58]. This method requires the specification of a “signature”, which lists all of the classes and properties that an application-specific use requires. The result of performing a modularization task is a sub-ontology for which all facts asserted or implicit in the source ontology, with regard to the signature classes, are still implied by the module.
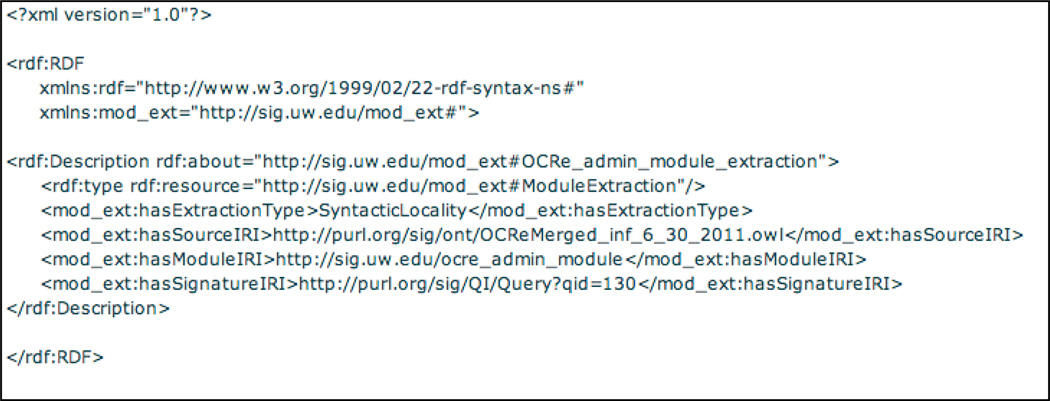
However, there is no standard declarative language for specifying a module extraction task. To address this deficit, we created a custom RDF schema, a configuration file format, for specifying module extraction “queries”. Query 203 (Figure 11) implements such a format for extracting the ocre_admin_module from the Ontology of Clinical Research (OCRe) [59], which defines the administrative elements for a clinical study. The signature for this extraction is supplied by a separate vSPARQL query 130 over OCRe (not shown) that is executed by the QES, thus eliminating the need to manually specify the signature as in current module extraction methods, and illustrating that QI can combine the strengths of both the graph-based and logical approaches in order to generate ontology views. The results of the signature query specify only the following classes out of several hundred, as starting points for extraction of the admin module: Physical entity, Person, Study site, Telecommunication address, Study, Organization, and Address.
Figure 11.
Query 203, module extraction input “schema”, which specifies such properties as the location of the input OWL ontology (OCRe), the type of extraction to perform (Syntactic Locality) and the signature, which is generated by the QES via query 130 (not shown).
4.2. Value set extraction for use in Electronic Data Capture (queries 130, 132, 135, 197, 198 and 203)
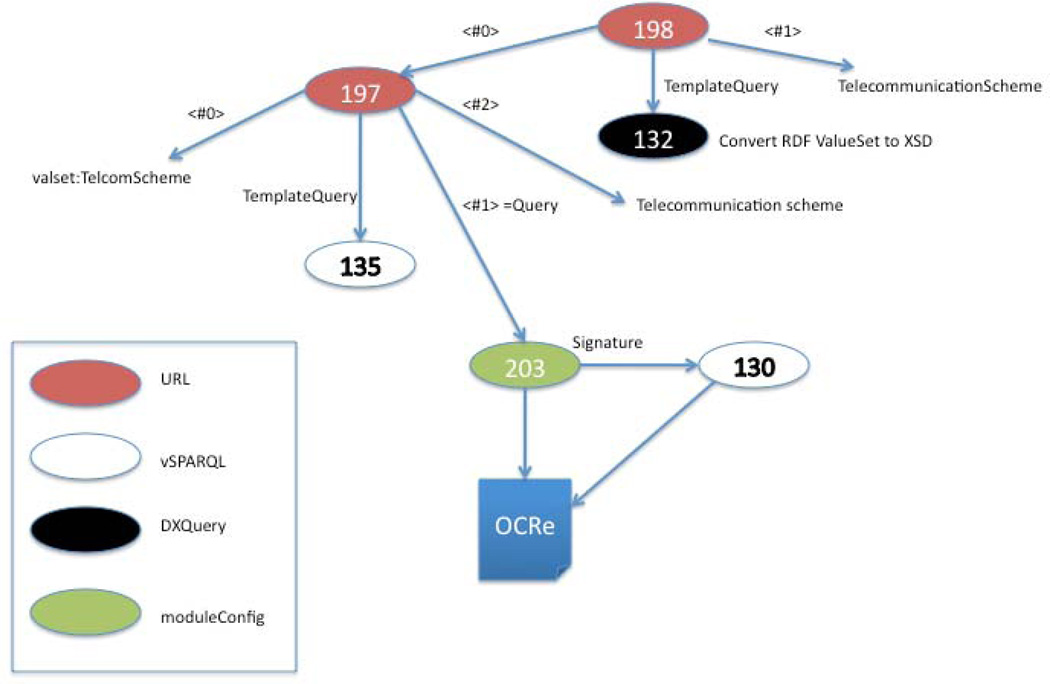
Figure 12 illustrates the generation of RDF value sets from the OCRe module extracted by query 203 described in the last section (Figure 11), and then transformation of the value set list to XML Schema (XSD) for use by an Electronic Data Capture (EDC) application, all done on-the-fly [60]. Value sets are used in clinical research to constrain the possible values that can be assigned to a data element on a case report form used in an EDC system. In this case the value set is the possible values for Telecommunication Scheme, which is one potential data element in the description of a clinical trial that will be recorded as part of the Human Studies Database (HSDB) [24].
Figure 12.
Chain of 6 queries that together dynamically generate both an RDF (query 197) and XSD (query 198) value set (Telecommunication scheme) from an extracted module of OCRe (query 203) based on a signature specified in query 130. Four different query “languages” are linked in this small query web.
Query 198, which results in the XSD value set, is one of a chain of 6 queries:
130 is a vSPARQL query that constructs the signature for 203
203 is a moduleConfig query that extracts a module from OCRe
135 is a vSPARQL template query that finds all instances of a given class from an ontology given as argument <#1>
197 is a URL query that sets the arguments for 135, in particular the extracted module generated by query 203 is assigned to argument <#1> (the source for query 197) and “Telecommunication_scheme” is assigned to argument <#2> (the root class label).
132 is a DXQuery template query that converts an RDF value set description provided by argument <#0> to XSD
198 sets the arguments for 132, in particular argument <#0>, which is the RDF value set generated by 197.
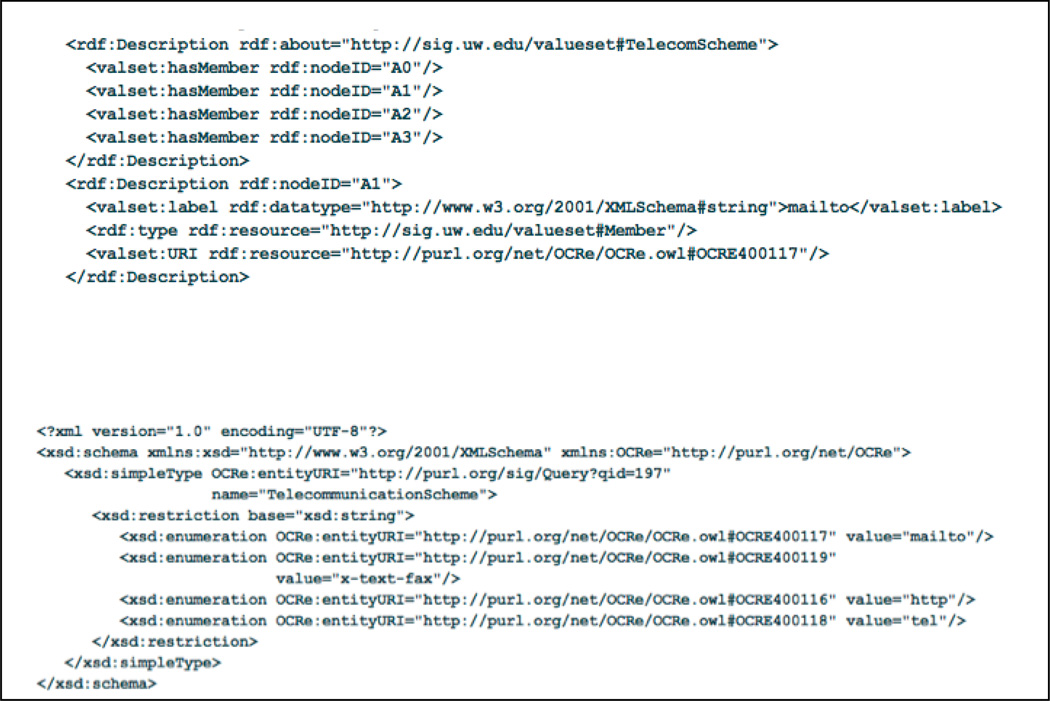
The result of executing query 197 is the desired value set in RDF; the result of executing 198 is the desired value set in XSD (Figure 13). These results could either be imported into an EDC application like REDCap [1], or could be dynamically accessed by a data annotation or data integration application.
Figure 13.
Top. Portion of telecommunication value set in RDF format, as generated by query 197. Bottom. Telecomunication value set in XSD format, as generated by query 198, based on the results from query 197.
4.3. Image annotation using an ontology-based terminology service (query 95)
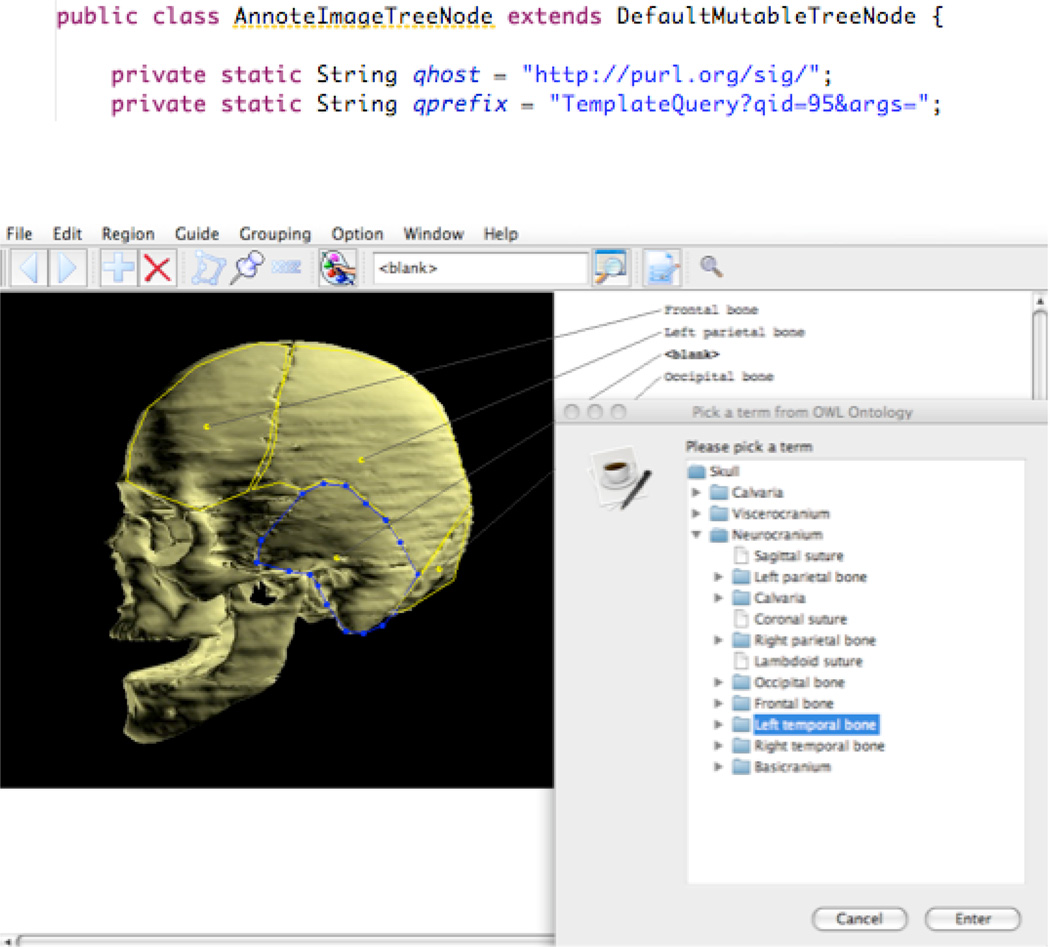
To illustrate the use of dynamic value sets for image annotation, consider template query 95, which finds the direct parts of an anatomical structure from the FMA. Figure 14 (top) shows a call to this query embedded in part of the Java code for our AnnoteImage application [61], and Figure 14 (bottom) shows the user interface of AnnoteImage. The user has outlined a structure on an image, and a call to the saved query has initially retrieved the top-level parts of the Skull (Calvaria, Viscerocranium and Neurocranium). The user then clicked on one of these parts (Neurocranium), which caused the template query to be re-run with Neurocranium as parameter, returning the parts of the Neurocranium. While the user interface presents a Windows Explorer style navigational tree, underneath it is calling the template query multiple times with different parameters.
Figure 14.
Use of a value set template query to provide a controlled list of terms for annotating regions of interest on an image. Top) Relevant code embedded in the AnnoteImage application. In a later part of the code qhost and qprefix are concatenated with an anatomical term to generate a query to QI. Bottom) Resulting user interface.
4.4. Local integration of clinical research data (queries 93 and 202)
In our Integrated Brain Imaging Center (IBIC) [62] and Institute for Translational Health Sciences (ITHS – a CTSA awardee) [63], an approach to data management that we are exploring is to store different types of data in different, generally open source database systems, each of which is optimized for a specific kind of data. This approach is an alternative to more typical monolithic databases that in our experience tend to become bloated, difficult to maintain and evolve, and expensive. However, since in our approach the data are in separate databases we need to integrate them. Because most such database systems now provide some sort of web service interface (often REST), we have experimented with using QI to integrate data across these databases.
Query 93 is an example of integrating data captured by the REDCap EDC system [1] and the XNAT imaging system [2], in this case for quality control (QC) [64]. The query finds all images that have been marked in XNAT as being unusable, checks REDCap for the exam date, and returns a report specifying the time remaining to acquire a new image within a specified time window.
Query 202 is an example of integrating data captured by REDCap with data in our RLab freezer management system. In this case REDCap records observational clinical trials of patients with lupus, whereas RLab manages aliquots of blood obtained from these patients and stored in a lab freezer for subsequent analysis. The query asks the freezer management system whether there any unused aliquots for patients whose REDCap record shows that they still need qPCR [65].
Both these queries access simulated data (which are nevertheless based on ongoing studies) since QI does not yet deal with security.
4.5. Wide-scale integration of cancer clinical research data (query 157)
The same mechanisms we are using for local data integration are applicable to wider-scale data integration. For example, we have been involved in an NCI contract to use caBIG tools [20] for sharing data about prostate cancer biospecimens across nine participating prostate cancer SPORE sites [71]. Each of the participating institutions has a local data management system to keep track of their own prostate specimens and associated clinical data. For this project a common XML schema has been designed for each of the local databases to export to, after which the XML data are imported into caTissue [72] for subsequent sharing on caGRID [73]. For the initial phase of this project only non-identifiable “green” data are exported, so security is not an issue. However, experience to-date confirms the findings of a recent caBIG report [35], which found that the caBIG tools are extremely difficult to install and use, to the point that the project has essentially given up trying to import the XML files into caTissue.
To explore whether our lightweight QI approach could help we created a set of partially simulated XML files that correspond to the prostate-spore schema, made them available at five separate web sites under our control, and then created saved XQuery 157, which asks for the records for all men over the age of 69 that had a prostatectomy, returning 2551 out of a total 7066 records at the four sites (at the time of this writing). The query accesses a “registry.xml” file, which lists the URLs of all five sites, where each site in the registry has the (simulated) XML file as might be exported by a single institution. The parent query 157 uses the XQuery “collection” function to automatically treat all XML files listed by the registry as if they were a single source. Adding a new site is thus as simple as adding a single line to the registry.xml file function, no query modification is necessary. This query took only a few minutes to create, and executes in about 10 seconds.
4.6. Wide-scale integration of clinical research metadata via a single instance of QI (query 163)
A second example of the potential of our approach for wide-scale clinical data integration is the Human Studies Database Project (HSBD [24]), whose goal is to create a distributed database of information about clinical trials that will allow researchers and others to find trials of interest. Interoperability will be assured because each individual database will use the Ontology of Clinical Research (OCRe) [59] to provide common terms and relations. Although the HSDB project is exploring multiple approaches to storing the data, one current approach is to derive a common XML schema from OCRe and import existing data into this schema. The HSDB team has explored both caBIG and I2B2 [66] as possible data integration methods, but both seem to be very heavyweight. Thus, the group is currently using our QI as a lightweight method for querying across data stores.
As proof of concept for this approach several example XML files corresponding to a simple XML schema were created and saved at several web sites. Query 163 is an XQuery that queries across these sites to find all studies whose Study Design is of type “Interventional study design”. A complication for this query is that the XML data are annotated only with subclasses of type Interventional study design (i.e. Crossover study design), not Interventional study design itself. Thus, the query first consults OCRe via vSPARQL template query 164 to find subclasses of Interventional study design, then looks for studies whose Study Design is one of these subclasses, in a similar manner to query 188 for brain parts.
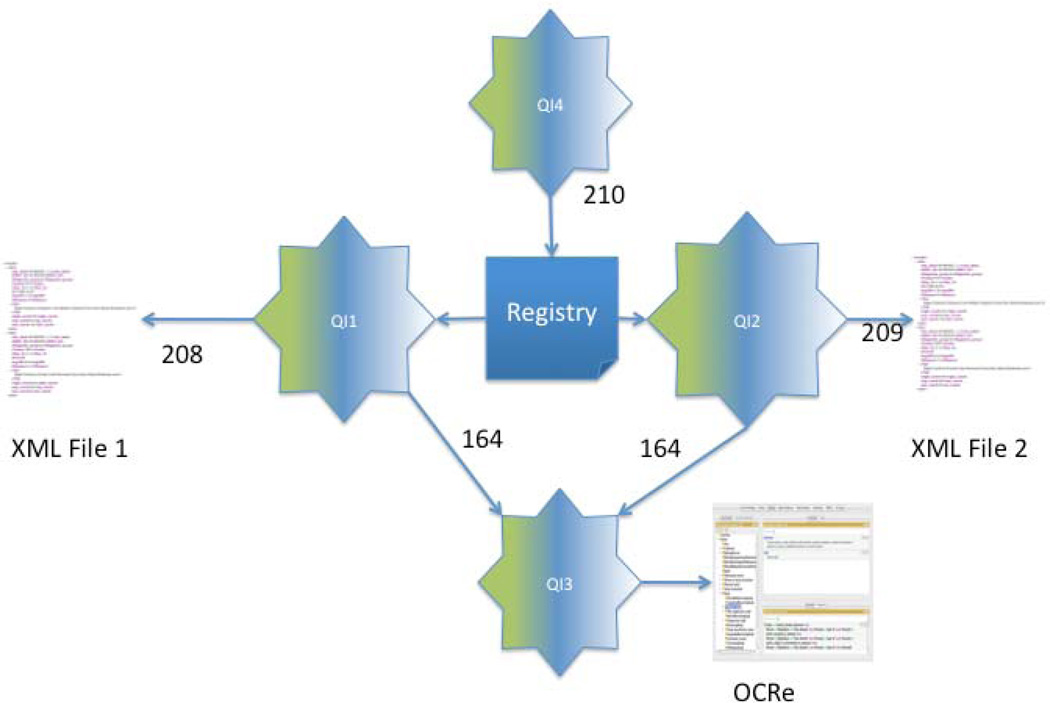
4.7. (Simulated) wide-scale integration of clinical research metadata via multiple instances of QI (queries 164, 208, 209 and 210)
A potential problem with the HSDB example described in the previous section is that the response time could become very slow if the XML files generated by each institution become large (which has so far not been a problem in the current instantiation of the HSDB). One reason for this potential problem is that each XML file is sent in its entirety from the source institution to the single instance of QI running in our lab (document shipping [67]), so network delays could be a major contributor to response time. One solution to this problem is to ship the query to the source (query shipping [67]) rather than shipping the document to the query engine. However, in order to make query-shipping work the source must have the capability of processing the query and returning the results.
Here we illustrate how query shipping could work by using multiple instances of QI, one at each source site. Although the queries execute, the scenario is simulated since each of the urls we use points to a single production installation of QI. We have not yet installed a second production version since we are still developing an easily installable version, but we have shown that our development server can access queries stored on our production version.
Figure 15 illustrates the simulation scenario we have implemented. Query 163 is replicated at each of two simulated instances of QI, QI1 (query 208) and QI2 (query 209). The query is slightly edited at each site such that the source XML file is the file generated at that site. In addition it is assumed that ontology query 164, which finds all subclasses of a type of study design, is located on a third instance QI3, which the queries at QI1 and QI2 access (perhaps a dedicated ontology views instance of QI that is eventually folded into Bioportal [45]). Yet a fourth simulated instance of QI, QI4, contains query 210, which runs a query over all the sources found in a registry.xml file. However, instead of recording the location of the XML files themselves as in query 163, the registry records the URLs of the queries 208 and 209 saved at each of QI1 and QI2. When each “source” is processed by the XQuery collection function in query 210, each of QI1 and QI2 executes its edited form of query 163 over its own XML file (consulting the centralized ontology at QI3 in the process), and then returns the results to QI4 where they are joined via the collection function. The result is that no XML data sources are sent over the network between institutions, only queries to each source and the results of running the queries at the sources.
Figure 15.
Illustration of a type of federated database using multiple instances of QI. Each of several institutions has its own instance of QI, each of which accesses its own XML file (or database) using the same saved query. Each local query in turn accesses a shared ontology web server implemented in a third instance of QI, and a fourth instance of QI joins the results from all the localized queries, there by removing the need to send large XML data sources over the network and allowing individual institutions to maintain control of their own data.
Many other possible configurations of this small query web can be envisioned since the ability to chain queries leads essentially to a series of query web subroutines that can be almost (or perhaps as) flexible as functional programming languages like LISP, since LISP syntax in particular has many similarities to XML and in fact was one of the inspirations for our design [68] (p. 208).
5. Discussion
In this paper we have described a Query Web Integrator and Manager (QI) that has the potential to tie together many of the currently fragmented web data and knowledge sources through the emergence of an interlinked web of queries. The key ideas that make this possible are the use of XML as the underlying data exchange mechanism, a framework that permits an agnostic view as to the best representation and query language on top of XML, the ability to save and re-use queries, and the ability to treat any saved query as a data source (a view) that can be queried by other queries, either in the same instance or in a separate instance of QI.
Within the context of the classification scheme provided by Goble [29] our approach can be primarily characterized as a lightweight view integration method, in that it combines the advantages of views over diverse data sources with the ability for anyone to easily create, edit and save those views such that they are reusable by others. Our approach also has characteristics of other data integration architectures, all within a lightweight framework. That is, a large database stored with a given instance of QI is like a central repository; a view over multiple sources materialized in a given instance of QI is like a warehouse; and distributed instances of QI all running the same query is one type of federated query system.
The ability to chain queries makes QI somewhat similar to workflow systems like Taverna [35], which allows users to graphically tie external web services together by connecting the output of one service to the input of another, and to save and execute the resulting workflow. Workflows in Taverna, which are represented as XML files, can be saved in a central repository called My Experiment [69], where they can re-used by others and included as part of larger workflows. Taverna also provides access to a large number of web services, including over 2200 services registered in the BioCatalogue central registry [70].
The Taverna toolkit is basically a way to glue together existing web services; unlike QI it does not in itself include methods for creating and saving queries. On the other hand, the availability of the QI REST interface means that queries saved in QI can be included within larger Taverna workflows that might, for example, provide graphical means for entering parameters for template queries, or provide additional processing on the results of queries over multiple sources. In turn, since Taverna workflows are represented in XML QI could potentially provide more sophisticated searching of Taverna workflows and/or available BioCatalogue web services through saved XQueries over the My Experiment or BioCatalogue REST search services.
QI at its current stage of development is not designed for the end-user, although motivated end-users have used it. Because query languages like SPARQL and XQuery are generally too arcane for most end-users, and because XML output is not easy to read, QI seems best suited at this point for experts in the queries and data sources, who will write and save queries for others to either click on within QI, or more likely, to access via workflows like Taverna or end-user applications like our AnnoteImage program (section 4.3). This scenario is not that different from current large-scale data management installations where experts are hired to write complex SQL reports, which are then simply clicked on by most users. An advantage of the Query Web approach is that these “reports” or “views”, in the form of saved queries, are available to anyone on the web who has authorization, thus potentially highly leveraging the skills of a few experts for a much larger end-user community.
Of course there are many substantial research issues that need to be addressed before QI can become more widely used. These issues include security, reliability, response time, discoverability, usability, ease of installation and customization among others. Many of these issues have been addressed by others in different contexts, and in future work we will explore the applicability of these methods to QI. However, even in its current state QI has already shown in our own work that it is useful for data integration problems in which security, discoverability and response times are not issues.
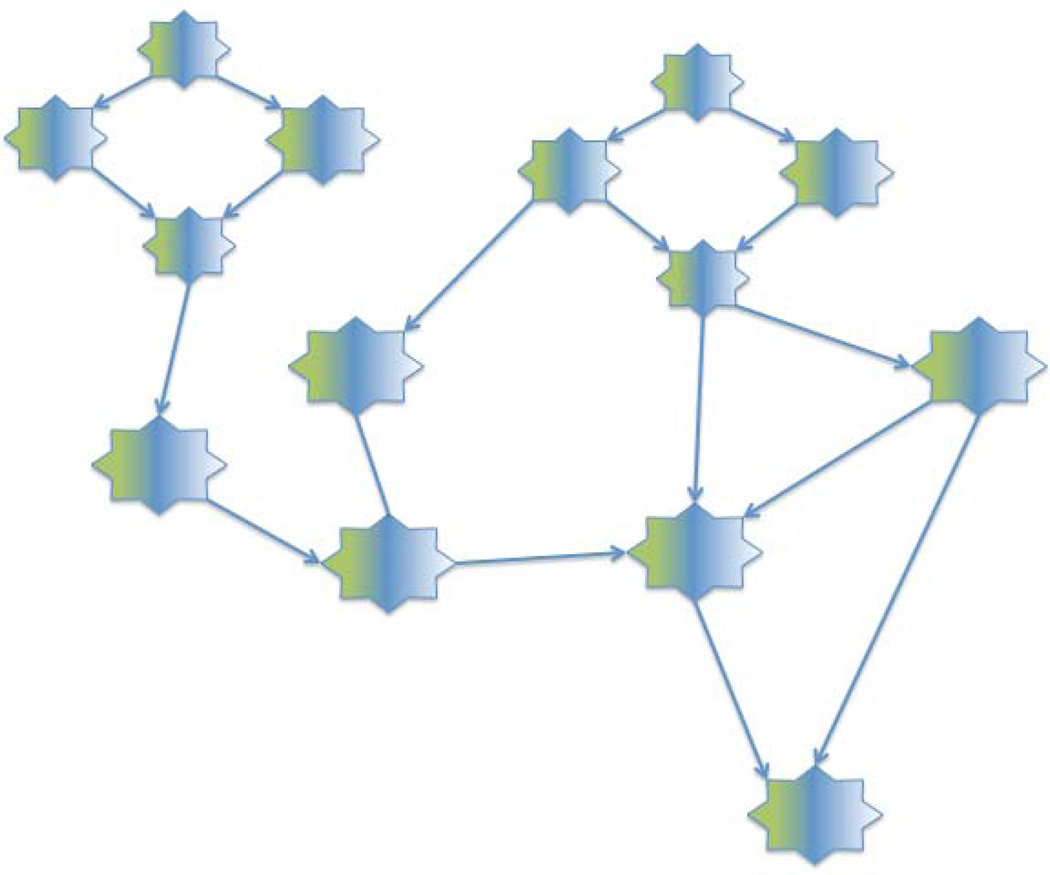
The fact that anyone can create queries, as views that can themselves be queried, leads to the potential evolution of a Query Web over the existing document and semantic web. Figure 3, Figure 12 and Figure 15 each illustrates a small query web, each of which could grow as users write queries that link to these chained queries. Figure 16 extends this idea, suggesting that users may want to integrate results from smaller query webs with the results from queries over other data and knowledge sources and other query webs. For example, as an extension of the queries generated in section 3 users may want to integrate genomic, clinical and imaging data in order to search for biomarkers of schizophrenia.
Figure 16.
As queries stored in separate instances of QI (as well as other query services) become chained together they could begin to evolve into a larger scale Query Web, which would be layered over the document web, and which would interoperate with the semantic web, linked data, and other web-accessible information resources.
In this way smaller query webs could gradually coalesce into the larger Query Web, with popular queries being accessed often, and less popular or even incorrect queries eventually dying out through disuse. Thus, like the document web and semantic web, the Query Web could evolve organically rather than through monolithic or arbitrary constraints that can never keep up with the rapidly changing nature of biology.
As the research issues become resolved QI should become increasingly useful for ever more diverse users and use cases. But even now we believe it could be of use for similar use cases to those described in this paper. We plan to have an open source downloadable version available soon (through the QI project page [49]). New installations, together with new services as well as end-user-centered interfaces, should not only be useful at the local level, but could also start to link to one another, thereby initiating the evolution of the Query Web. Such a Query Web has the potential to encompass much more than biomedicine since, like the Web itself, there is nothing in the technology that limits it to a single field.
Supplementary Material
Acknowledgements
This work was primarily funded by NIH grant HL08770, with additional funding for some of the specific queries by NIH grants RR025014, AR051545, RR026040, NS073008, and NCI contract S10-029. The work builds on the efforts of many individuals over the years, for development of many of the information sources and ontologies, program code, and queries that are currently saved in QI. Specifically:
Cornelius Rosse and Jose L.V. Mejino: Foundational Model of Anatomy
Dan Cook: Ontology of Physics for Biology
Marianne Shaw and Dan Suciu: vSPARQL
Nicola Dell, Linda Shapiro: VIQUEN
Chris Re, Nathan Bales, Eider Moore and Dan Suciu: Distributed XQuery
Eider Moore: Mind Seer
Jessica Turner: BIRN fMRI data
Nolan Nichols, Marianne Shaw, Xenia Hertzenberg, and Ron Shaker wrote many of the saved queries as described in this paper. Specific authors are shown in the list of queries accessible via the project web site [49].
We also greatly profited from our discussions with Natasha Noy and Mark Musen during our collaborative RO1 with the National Center for Biomedical Ontology (the RO1 which funded this work); Jessica Turner and Daniel Rubin during our collaboration on the RadLex project and neuroscience data integration; Ida Sim, Samson Tu and Simona Carini during our collaboration on the Human Studies Database Project, and Marianne Martone and Jeff Grethe of the Neuroscience Information Framework.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability
We are currently working to create an easily installable and open source version of the Query Integrator, which will be available from the QI project page at http://si.washington.edu/projects/QI. In the meantime we are happy to work with knowledgeable developers who are interested in experimenting with and perhaps contributing to prototypes of this system. Please email the corresponding author if you are interested in this possibility.
Contributor Information
James F. Brinkley, Email: brinkley@u.washington.edu.
Landon T. Detwiler, Email: det@u.washington.edu.
REFERENCES
- 1.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18929686 18929686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcus DS, Olsen T, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit (XNAT): An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. http://docs.xnat.org/file/view/xnat_neuroinformatics.pdf/229736086/xnat_neuroinformatics.pdf. [DOI] [PubMed] [Google Scholar]
- 3.Jakobovits RM, Rosse C, Brinkley JF. WIRM: An open source toolkit for building biomedical web applications. J Am Med Ass. 2002;9(6):557–590. doi: 10.1197/jamia.M1138. http://sigpubs.biostr.washington.edu/archive/00000134/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong C, Brinkley J. Customizable Electronic Laboratory Online (CELO): A web-based data management system builder for biomedical research laboratories; Proceedings, Fall Symposium of the Amercian Medical Informatics Association; 2006. p. 922. http://sigpubs.biostr.washington.edu/archive/00000191/ [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY, Cochrane GR. The 2011 Nucleic Acids Research database issue and online molecular biology database collection. Nucleic Acids Research. 2011;39(Supplement 1):D1–D6. doi: 10.1093/nar/gkq1243. http://nar.oxfordjournals.org/content/39/suppl_1/D7.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. (Database issue) http://nar.oxfordjournals.org/cgi/reprint/35/suppl_1/D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Center for Biotechnology Information. Home Page. 2011 http://www.ncbi.nlm.nih.gov.
- 8.Flybase. A Database of Drosophila Genes and Genomes. 2011 doi: 10.1007/978-1-4939-6371-3_1. http://flybase.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zfin. The Zebrafish Model Organism Database. 2011 http://zfin.org. [Google Scholar]
- 10.The Jackson Laboratory. Mouse Genome Informatics. 2011 http://www.informatics.jax.org/. [Google Scholar]
- 11.KEGG. KEGG: Kyoto Encyclopedia of Genes and Genomes. 2005 http://www.genome.ad.jp/kegg. [Google Scholar]
- 12.Karp P. Pathway databases: a case study in computational symbolic theories. Science. 2001;293(2040–2044) doi: 10.1126/science.1064621. [DOI] [PubMed] [Google Scholar]
- 13.OpenMRS. Open Medical Record System. 2011 http://openmrs.org. [Google Scholar]
- 14.Microsoft. Amalga. 2011 http://www.microsoft.com/enus/microsofthealth/products/microsoft-amalga.aspx. [Google Scholar]
- 15.Akaza Research. OpenClinica. 2011 http://openclinica.com. [Google Scholar]
- 16.Medidata. Rave. 2011 http://www.mdsol.com/products/rave_overview.htm. [Google Scholar]
- 17.Velos. eResearch. 2011 http://www.velos.com/products_eres_overview.shtml. [Google Scholar]
- 18.Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro JP, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) J Am Med Inform Assoc. 2011;18(4):416–422. doi: 10.1136/amiajnl-2010-000032. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21515543 21515543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardiovascular Research Grid (CVRG) Home Page. 2010 http://cvrgrid.org/. [Google Scholar]
- 20.National Cancer Institute. caBIG Home Page. 2010 https://cabig.nci.nih.gov/.
- 21.Hochheiser H, Aronow BJ, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, Fraser SE, Iwata J, Marazita ML, Murray JC, Murray S, Postlethwait J, Potter S, Shapiro LG, Spritz R, Visel A, Weinberg SM, Trainor PA. The FaceBase Consortium: A comprehensive program to facilitate craniofacial research. Developmental Biology. 2011 doi: 10.1016/j.ydbio.2011.02.033. In Press http://dx.doi.org/10.1016/j.ydbio.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GenitoUrinary Development Molecular Anatomy Project (GUDMAP) Home Page. 2010 http://www.gudmap.org/. [Google Scholar]
- 23.Neuroscience Information Framework (NIF) Home Page. 2010 http://nif.nih.gov/.
- 24.Sim I. The Human Studies Database (HSDB) 2010 http://rctbank.ucsf.edu/home/hsdb.html. [PMC free article] [PubMed] [Google Scholar]
- 25.Center for Disease Control. WONDER. 2011 http://wonder.cdc.gov/.
- 26.Stein L. Creating a bioinformatics nation. Nature. 2002;417:119–120. doi: 10.1038/417119a. [DOI] [PubMed] [Google Scholar]
- 27.Bizer C, Heath T, Berner Linked data - the story so far. International Journal on Semantic Web and Information Systems (IJSWIS) 2009;5(3):1–22. http://tomheath.com/papers/bizer-heath-berners-lee-ijswis-linked-data.pdf. [Google Scholar]
- 28.JSON. Introducing JSON. 2012 http://www.json.org/. [Google Scholar]
- 29.Goble C, Stevens R. State of the nation in data integration for bioinformatics. Journal of Biomedical Informatics. 2008;41:687–693. doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Halevy AY. Answering queries using views: a survey. VLDB J. 2001;10(4):270–294. [Google Scholar]
- 31.Baker PG, Goble CA, Bechhofer S, Paton NW, Stevens R, Brass A. An ontology for bioinformatics applications. Bioinformatics. 1999;15(6):510–520. doi: 10.1093/bioinformatics/15.6.510. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10383475 10383475. [DOI] [PubMed] [Google Scholar]
- 32.Shaker R, Mork P, Brockenbrough JS, Donelson L, Tarczy-Hornoch P. The Biomediator system as a tool for integrating biologic databases on the web. Proc. Workshop on Information Integration on the Web, held in conjunction with VLDB. 2004 http://www.biomediator.org/publications/iiweb-paper-biomediator.pdf. [Google Scholar]
- 33.Gupta A, Bug W, Marenco L, Qian X, Condit C, Rangarajan A, Muller H, Miller P, Sanders B, Grethe JS, Astakhov V, Shepherd G, Sternberg PW, Martone ME. Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF) Neuroinformatics. 2008;6:205–217. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.JAQL. Query language for JavaScript object notation. 2012 http://code.google.com/p/jaql/. [Google Scholar]
- 35.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, Li P. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20(17):3045–3054. doi: 10.1093/bioinformatics/bth361. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15201187 15201187. [DOI] [PubMed] [Google Scholar]
- 36.PostgreSQL Global Development Group. PostgreSQL. 2011 http://www.postgresql.org/. [Google Scholar]
- 37.Dell NL, Shapiro LG, brinkley JF. VIQUEN: A visual query engine for RDF. 2011 Submitted. [Google Scholar]
- 38.Re C, Brinkley J, Hinshaw K, Suciu D. Distributed XQuery. Proceedings of the Workshop on Information Integration on the Web (IIWeb) 2004:116–121. http://sigpubs.biostr.washington.edu/archive/00000157/ [Google Scholar]
- 39.Detwiler LT, Suciu D, Franklin JD, Moore EB, Poliakov AV, Lee ES, Corina D, Ojemann GA, Brinkley JF. Distributed XQuery-based integration and visualization of multimodality data: Application to brain mapping. Frontiers in Neuroinformatics. 2009;3(2) doi: 10.3389/neuro.11.002.2009. http://sigpubs.biostr.washington.edu/archive/00000234/ PMCID: PMC2636687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw MS, Detwiler LT, Noy NF, Brinkley JF, Suciu D. vSPARQL: A view definition language for the semantic web. Journal of Biomedical Informatics. 2010;44(1):102–117. doi: 10.1016/j.jbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.HP Labs. Jena - A semantic web framework for Java. 2011 http://www.openjena.org. [Google Scholar]
- 42.Rosse C, Mejino JLV. A reference ontology for bioinformatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. http://www.sciencedirect.com/science/article/pii/S1532046403001278. [DOI] [PubMed] [Google Scholar]
- 43.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI thesaurus. Methods of Information in Medicine. 2005;44:498–507. http://sig.biostr.washington.edu/share/sigweb/pubs/SmithNCITanalysis.pdf. [PubMed] [Google Scholar]
- 44.Reactome Group. Reactome. 2011 http://www.reactome.org. [Google Scholar]
- 45.National Center for Biomedical Ontology (NCBO) BioPortal. 2010 http://bioportal.bioontology.org/.
- 46.Shaw M. View definition languages for Biomedical RDF Ontologies. Seattle: University of Wasington; 2011. [Google Scholar]
- 47.Apache Software Foundation. Apache Tomcat. 2007 http://tomcat.apache.org/. [Google Scholar]
- 48.Moore EB, Poliakov A, Lincoln P, Brinkley J. MindSeer: A portable and extensible tool for visualization of structural and functional neuroimaging data. BMC Bioinformatics. 2007;8:389. doi: 10.1186/1471-2105-8-389. http://www.biomedcentral.com/1471-2105/8/389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Structural Informatics Group. Query Web Integrator and Manager (QI) 2011 doi: 10.1016/j.jbi.2012.03.008. http://si.washington.edu/projects/QI. [DOI] [PMC free article] [PubMed]
- 50.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers; 1988. [Google Scholar]
- 51.Potkin SG, Turner JA, Brown GG, McCarthy G, Greve DN, Glover GH, Manoach DS, Belger A, Diaz M, Wible CG, Ford JM, Mathalon DH, Gollub R, Lauriello J, O'Leary D, van Erp TG, Toga AW, Preda A, Lim KO. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009;35(1):19–31. doi: 10.1093/schbul/sbn162. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19042912 19042912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner JA, Mejino JLV, Brinkley JF, Detwiler LT, Lee HJ, Martone ME, Rubin DL. Applicaton of neuroanatomical ontologies for neuroimaging data annotation. Frontiers in Neuroinformatics. 2010;4(10) doi: 10.3389/fninf.2010.00010. http://www.frontiersin.org/neuroinformatics/10.3389/fninf.2010.00010/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brinkley JF, Turner J, Detwiler LT, Mejino JL, Martone M, Rubin D. Intelligent queries over BIRN data using the Foundational Model of Anatomy and a distributed query-based data integration system; Proceedings AMIA Fall Symposium; 2010. p. 989. http://sigpubs.biostr.washington.edu/archive/00000248/ [Google Scholar]
- 54.Brett M. The MNI brain and the Talairach atlas. 2006 http://imaging.mrc-cbu.cam.ac.uk/imaging/MniTalairach. [Google Scholar]
- 55.Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10(3):120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. http://ric.uthscsa.edu/projects/talairachdaemon.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3D statistical neuroanatomical models from 305 MRI volumes; Proceedings, IEEE Nuclear Science Symposium and Medical Imaging Conference; 1993. pp. 1813–1817. [Google Scholar]
- 57.Neuroimaging in Python Team. Nipype: Neuroimaging in Python pipelines and interfaces. 2011 http://nipy.sourceforge.net/nipype/. [Google Scholar]
- 58.Pathak J, Johnson TM, Chute CG. Survey of modular ontology techniques and their applications in the biomedical domain. Integrated Computer-Aided Engineering. 2009;16:225–242. doi: 10.3233/ICA-2009-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carini S, Harris S, MacCallum P, Rector A, Sim I, Toujilov I, Tu S. The Ontology of Clinical Research (OCRe) 2009 http://rctbank.ucsf.edu/home/ocre.html. [Google Scholar]
- 60.Brinkley JF, Detwiler LT. Query chains for dynamic generation of value sets; Proceedings, Fall Symposium of the American Medical Informatics Association; 2011. p. 1699. [Google Scholar]
- 61.Lober WB, Brinkley JF. A portable image annotation tool for web-based anatomy atlases; Proceedings, American Medical Informatics Association Fall Symposium; Washington, D.C. 1999. p. 1108. [Google Scholar]
- 62.Integrated Brain Imaging Center (IBIC) Home Page. 2011 http://www.ibic.washington.edu/. [Google Scholar]
- 63.University of Washington. Institute of Translational Health Sciences. 2008. http://iths.org/. [Google Scholar]
- 64.Nichols N, Detwiler LT, Franklin JD, Brinkley JF. Distributed queries for quality control checks in clinical trials; Proceedings, AMIA Spring Summit on Clinical Research Informatics; San Francisco. 2011. p. 115. http://sigpubs.biostr.washington.edu/archive/00000252/ [Google Scholar]
- 65.Wikipedia. Real-time polymerase chain reaction. 2011 http://en.wikipedia.org/wiki/Real-time_polymerase_chain_reaction. [Google Scholar]
- 66.I2B2. Informatics for Integrating Biology and the Bedside. 2011 https://www.i2b2.org/. [Google Scholar]
- 67.Ozsu MT, Valduriez P. Principles of Distributed Database Systems. New York: Springer; 2011. [Google Scholar]
- 68.Brinkley JF, Altman RB, Duncan BS, Buchanan BG, Jardetzky O. Heuristic refinement method for the derivation of protein solution structures: validation on cytochrome b562. J. Chem. Inf. Comput. Sci. 1988;28(4):194–210. doi: 10.1021/ci00060a005. http://sigpubs.biostr.washington.edu/archive/00000015/ [DOI] [PubMed] [Google Scholar]
- 69.De Roure D, Goble C. myExperiment. 2011 http://www.myexperiment.org/. [Google Scholar]
- 70.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Aleksejevs S, Stevens R, Pettifer S, Lopez R, Goble CA. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 38:W689–W694. doi: 10.1093/nar/gkq394. (Web Server issue) http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20484378 20484378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.