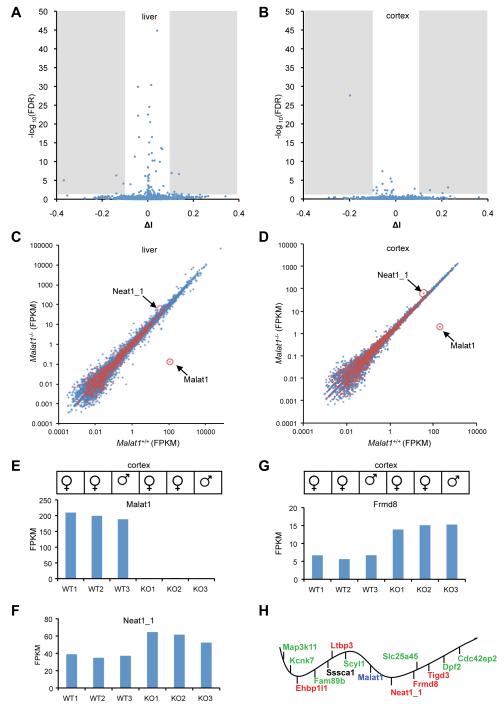
Figure 5. Malat1 RNA does not regulate global pre-mRNA splicing, but its transcription inactivation alters gene expression in cis.
A,B. The volcano plot shows proportional change of inclusion level (x axis, ΔI) of each exon and their statistical significance (y axis, -log10(FDR)) upon Malat1 depletion. Shaded regions represent statistical significant changes. C,D. The scatter plot shows the average expression of protein-coding (blue) and lncRNA (red) genes in wild type and Malat1 mutant livers and brain cortices. Malat1 and Neat1_1 are highlighted. E. Malat1 is significantly depleted in Malat1 mutant brain cortexes. F, G, Neat1_1 and Frmd8 show consistent overexpression in mutant cortexes. WT, wild type (Malat1+/+); KO, knockout (Malat1−/−). H. The ~240 kb genomic locus of Malat1 with adjacent genes distributed in order but not at the exact scale. Upregulated genes with statistical significance after multiple testing correction (red), and with statistical significance without multiple testing correction (green). Sssca1 expression is not significantly altered. Vertical bars under the wavy line represent transcription from the negative strand of the chromosome; vertical bars above the line represent transcription from the positive strand.