Table 1.
Binding affinities of adenosine derivatives at human A1, A2A, and A3ARs expressed in CHO cells (expressed as Ki value or percent displacement at 10 (M) and maximal agonist effects at 10 (M (% of full agonist) at the ARs
| Compound | 2-Substitution |
Ki at A1AR (nMa) (% activation) |
Ki at A2AAR (nMa) (% activation) |
% activation at A2BARa (and % inhibition), unless noted |
Ki at A3AR (nMa) (% activation) |
|---|---|---|---|---|---|
| 2-Alkoxy analogues | |||||
| 1 | CH3O | 155 ± 32 (119) | 970 ± 310 (78.6) | −2.7 ± 5.3 (−0.2) | 156 ± 37 (75.2 ± 5.1) |
| 2 | CH3CH2O | 2640 ± 540 (81.3) | 360 ± 139 (92.4) | −0.8 ± 1.9 (−6.7) | 568 ± 205 (99.1 ± 4.2) |
| 3 | (CH3)2CHO | 16% (15.9) | 927 ± 204 (87.5) | 0.4 ± 2.9 (−3.2) | 457 ± 154 (101 ± 5) |
| 4 | (CH3)2CHCH2O | 4410 ± 1150 (44.6) | 222 ± 89 (93.2) | −1.2 ± 8.8 (−3.4) | 84.2 ± 8.4 (108 ± 10) |
| 5 | (CH3CH2)2CHCH2O | 42% (1.0) | 45.8 ± 22.1 (90.0) | −4.4 ± 4.2 (2.0) | 336 ± 116 (4.9 ± 5.2) |
| 6 | Cyclohexyl-CH2O | 3350 ± 390 (27.5) | 342 ± 22 (92.8) | −0.9 ± 2.3 (−4.7) | 143 ± 32 (27.1 ± 4.9) |
| 7 | (CH3)2CH(CH2)2O | 3560 ± 1120 (29) | 37.7 ± 4.9 (92.4) | −0.6 ± 1.8 (−8.4) | 81.1 ± 9.0 (96.3 ± 3.0) |
| 8 | Cyclohexyl-(CH2)2O | 36% (27.8) | 579 ± 250 (102) | −4.6 ± 8.8 (0.4) | 578 ± 182 (51.6 ± 3.4) |
| 9 | 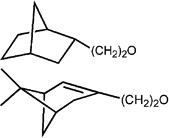 |
3590 ± 670 (0) | 137 ± 31 (98.6) | −0.1 ± 2.6 (2.8) | 149 ± 45 (−3.5 ± 9.0) |
| 10 | 3350 ± 720 (25.9) | 21.2 ± 12.7 (97.7) | −0.5 ± 4.4 (2.8) | 341 ± 132 (−0.2 ± 5.9) | |
| 11 | (CH3)2CH(CH2)3O | 3700 ± 790 (18.3) | 77.8 ± 20.9 (96.4) | −1.7 ± 2.8 (0.0) | 105 ± 31 (49.6 ± 8.1) |
| 12 | Cyclohexyl-(CH2)3O | 1730 ± 330 (33.5) | 92.0 ± 52.5 (105) | −2.5 ± 4.6 (2.2) | 83.3 ± 8.4 (21.2 ± 11.8) |
| 13 | Cyclohexyl-(CH2)4O | 883 ± 99 (109) | 291 ± 73 (98.5) | −0.27 ± 2.8 (−0.5) | 105 ± 13 (12.7 ± 1.5) |
| 14 | CH3(CH2)4O | 2430 ± 620 (48.7) | 6.9 ± 1.3 (94.1) | −2.2 ± 2.6 (9.9) | 222 ± 68 (92.8 ± 13.4) |
| 15 | CH3C≡C–(CH2)2O | 583 ± 78 (49.8) | 63.2 ± 50.3 (95.9) | 1.4 ± 3.7 (−7.9) | 90.2 ± 36.2 (73.4 ± 4.4) |
| 16 | CH3(CH2)5O | 1830 ± 340 (11.8) | 156 ± 89 (100) | 12.6 ± 0.5 (−6.5) | 124 ± 28 (43.7 ± 6.3) |
| 17 | (CH3)2C=CH(CH2)2CH(R-CH3)(CH2)2O | 34% (23.6) | 382 ± 100 (91.3) | 0.3 ± 3.4 (−5.0) | 431 ± 130 (−1.5 ± 1.5) |
| 18 | (CH3)2C=CH(CH2)2CH(S-CH3)(CH2)2O | 4550 ± 950 (21.3) | 78.3 ± 10.3 (90.2) | −0.4 ± 1.7 (−1.7) | 74.4 ± 22.5 (31.5 ± 6.4) |
| 2-Aryl- and arylalkyloxo (or amino) | |||||
| 19 | Phenyl-O | 5140 ± 1110 (57.6) | 44 ± 12 (100) | 0.7 ± 1.8 (−0.9) | 364 ± 96 (32.2 ± 3.5) |
| 20 | Benzyl-O | 642 ± 79 (11.5) | 585 ± 155 (85.1) | −3.3 ± 0.6 (9.5) | 117 ± 8 (16.9 ± 3.9) |
| 21 | 3-Chlorobenzyl-O | 27.4 ± 3.9 (46) | 228 ± 66 | 7.4 ± 2.1 | 71.6 ± 24.6 (16.2 ± 3.8) |
| 22 | Phenyl-(CH2)2O | 221 ± 57 (112) | 9.3 ± 2.9 (99.6) | 3490 ± 1490c (0.0) | 54.2 ± 14.3 (70.7 ± 2.7) |
| 23 | Phenyl-(CH2)2NH | 530 ± 88 (70.5) | 62.0 ± 17.6 (105) | −1.7 ± 3.1 (−6.9) | 310 ± 163 (72.0 ± 3.2) |
| 24 | Phenyl-(CH2)2S | 3700 ± 770 | 590 ± 260 | ND | 1960 ± 310 |
| 25 | 2-Methylphenyl-(CH2)2O | 396 ± 83 (74.6) | 17.4 ± 7.4 (110) | −1.0 ± 2.7 (3.8) | 214 ± 47 (7.3 ± 6.0) |
| 26 | 3-Methylphenyl-(CH2)2O | 295 ± 8 (106) | 41.6 ± 22.0 (98.4) | 15.3 ± 2.5 (0.6) | 242 ± 55 (70.9 ± 3.9) |
| 27 | 4-Methylphenyl-(CH2)2O | 1250 ± 250 (61.8) | 118 ± 95 (98.3) | 12.1 ± 1.9 (−12.2) | 470 ± 81 (95.5 ± 15.1) |
| 28 | 2-Methyloxyphenyl-(CH2)2O | 490 ± 114 (111) | 274 ± 142 (94.2) | 7.4 ± 1.3 (0.6) | 940 ± 354 (3.6 ± 6.7) |
| 29 | 3-Methyloxyphenyl-(CH2)2O | 246 ± 34 (114) | 32.1 ± 1.6 (103) | 0.4 ± 4.1 (0.0) | 231 ± 53 (85.1 ± 5.8) |
| 30 | 4-Methyloxyphenyl-(CH2)2O | 288 ± 22 (109) | 64.3 ± 7.8 (97.6) | 11.3 ± 1.8 (−4.4) | 105 ± 20 (91.1 ± 9.2) |
| 31 | 3,4-Dimethyloxyphenyl-(CH2)2O | 469 ± 118 (101) | 30.3 ± 2.8 (99.8) | −1.4 ± 2.1 (6.2) | 863 ± 313 (66.5 ± 3.7) |
| 32 | 2-Fluorophenyl-(CH2)2O | 331 ± 22 (18.9) | 58.1 ± 24.9 (99.1) | 16.6 ± 2.5 (0.3) | 77.8 ± 13.5 (44.5 ± 5.1) |
| 33 | 4-Fluorophenyl-(CH2)2O | 467 ± 100 (115) | 56.8 ± 16.3 (97.9) | 17.3 ± 3.1 (−7.2) | 112 ± 16 (73.4 ± 5.2) |
| 34 | 2-Chlorophenyl-(CH2)2O | 366 ± 33 (31.8) | 17.9 ± 6.1 (94.9) | 1.0 ± 3.4 (6.9) | 144 ± 22 (1.4 ± 2.7) |
| 35 | 3-Chlorophenyl-(CH2)2O | 372 ± 116 (85.3) | 11.5 ± 5.3 (96.7) | 28.0 ± 4.9 (2.9) | 41.0 ± 7.8 (31.0 ± 7.0) |
| 36 | 4-Chlorophenyl-(CH2)2O | 331 ± 51 (113) | 58.5 ± 8.0 (97.4) | 17.5 ± 2.8 (5.7) | 116 ± 23 (69.0 ± 6.3) |
| 37 | R-2-Phenylbutyl-O | 28% (24.5) | 503 ± 98 (97.7) | −0.2 ± 6.0 (4.6) | 201 ± 61 (−1.6 ± 3.8) |
| 38 | S-2-Phenylbutyl-O | 4780 ± 990 (16.0) | 26.9 ± 6.9 (96.0) | 0.4 ± 5.6 (−5.5) | 175 ± 31 (−3.9 ± 3.9) |
| 39 | 2-(1-Naphthyl)ethyl-O | 220 ± 18 (101) | 3.8 ± 1.4 (102) | −2.1 ± 5.1 (−3.7) | 205 ± 19 (12.8 ± 5.9) |
| 40 | 2-(2-Naphthyl)ethyl-O | 141 ± 51 (102) | 16.1 ± 7.0 (105) | 1440 ± 70c | 130 ± 8 (45.1 ± 8.5) |
| 41 | 2,2-Diphenylethyl-O | 39% (10.4) | 310 ± 119 (97.5) | −1.8 ± 5.6 (0.8) | 53.6 ± 10.4 (−0.0 ± 0.6) |
| 42 | 2-(2-Thienyl)ethyl-O | 174 ± 20 (112) | 10.9 ± 4.8 (105) | 1780 ± 260c | 93.3 ± 16.8 (79.7 ± 4.5) |
| 43 | 2-(3-Thienyl)ethyl-O | 280 ± 72 (117) | 13.3 ± 4.1 (106) | 8.9 ± 5.5 (−2.0) | 101 ± 34 (61.6 ± 15.1) |
| 44 | trans-2-Phenylcyclopropyl-O | 367 ± 27 (19.1) | 2050 ± 700 | 1.2 ± 2.3 | 292 ± 46 (0.4 ± 1.6) |
| 45 | 4-Phenylbutyl-O | 1100 ± 230 (24.6) | 243 ± 166 (103) | −0.4 ± 2.3 (−7.8) | 251 ± 80 (19.3 ± 6.2) |
| 46 | 5-Phenylpentyl-O | 700 ± 126 (104) | 249 ± 54 (101) | 6.2 ± 3.6 (5.4) | 429 ± 159 (9.8 ± 8.8) |
| 5′,8-Cyclo analogues | Name | ||||
| 47 | Cyclo-CPAb | 418 ± 62 (117) | 12% (11.7) | −0.6 (−2.9) | 25% (11.7 ± 0.3) |
| 48 | Cyclo-R-PIAb | 49% (26.3) | 18% (27.6) | 4.5 (−9.6) | 27% (1.7 ± 1.7) |
| Reference compounds | Name | ||||
| 49 | CPA | 1.8 ± 0.5 (112) | 820 ± 216 (98) | 8.6 ± 2.9 (0.0) | 72 ± 12 (99 ± 6) |
| 50 | R-PIA | 2.0 ± 0.3 (101) | 884 ± 188 (101) | 1680 ± 498c | 8.7 ± 0.9 (102 ± 6) |
| 51 | CGS21680 | 1570 ± 460 (99) | 8.8 ± 1.6 (100) | 4.7 ± 1.8 (0.0) | 114 ± 16 (98 ± 5) |
| 52 | NECA | 6.8 ± 2.4 (100) | 2.2 ± 0.6 (100) | 140 ± 19c (0.0) | 16.0 ± 5.4 (100) |
At the A2BAR, EC50 values or the percent stimulation at 1 (M (and in parentheses percent inhibition at 1 (M of the effects of 100 nM NECA) are shown.
All experiments were performed using adherent CHO cells stably transfected with cDNA encoding a human adenosine receptor. Percent activation of the human A1 A2A and A3AR was determined at 10 µM. Binding at A1, A2A and A3ARs was carried out as described in Section 2. The A1 and A2AAR activation results were expressed as the mean values from two separate experiments, while the A3AR activation results were from three separate experiments. The Ki and EC50 values from the present study are means ± S.E.M., N = 3–5.
Structures shown in Fig. 4; ND: not determined.
EC50 (nM) for activation of the A2BAR.
