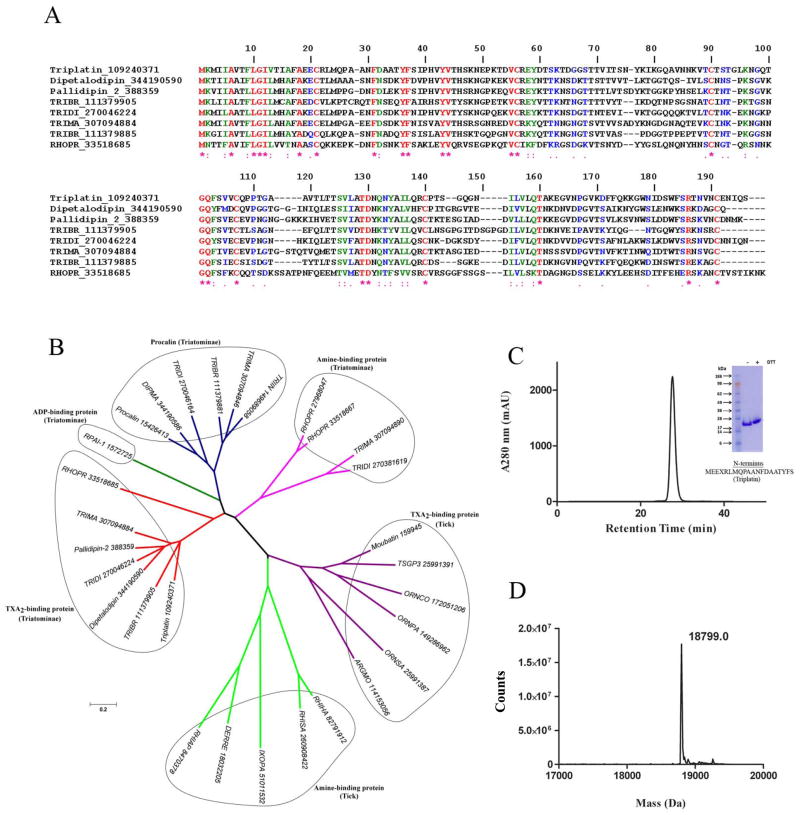
Fig. 1. Characterization of triplatin.
(A) Clustal alignment of triplatin (gi109240370) and DPTL (gi344190590), pallidipin-2 (gi388359), RPAI-1 (gi1572725), triabin (gi1122389) and several other triatomine putative TXA2-binding proteins. (B) Phylogenetic tree of triplatin and related salivary lipocalins. Tree was generated using the neighbor-joining method after 10,000 bootstraps. The numbers in the phylogram nodes indicates percent bootstrap support for the phylogeny. The bar at the bottom indicates 20% amino acid divergence in sequences. (C) Triplatin was loaded in a Sephadex G75 gel-filtration column and eluted at 1 ml/min in TBS, pH 7.4. Inset, gel electrophoresis of triplatin (reducing [+] and non-reducing [−] conditions) was carried out in a 4–12% NuPAGE gel. N-terminus was performed by Edman degradation. (D) Mass spectrometry of triplatin reveals a mass of 18,799 Da, which is in agreement with a theoretical mass of 18,672.7 Da with an extra methionine and 6xhis.