Figure 1.
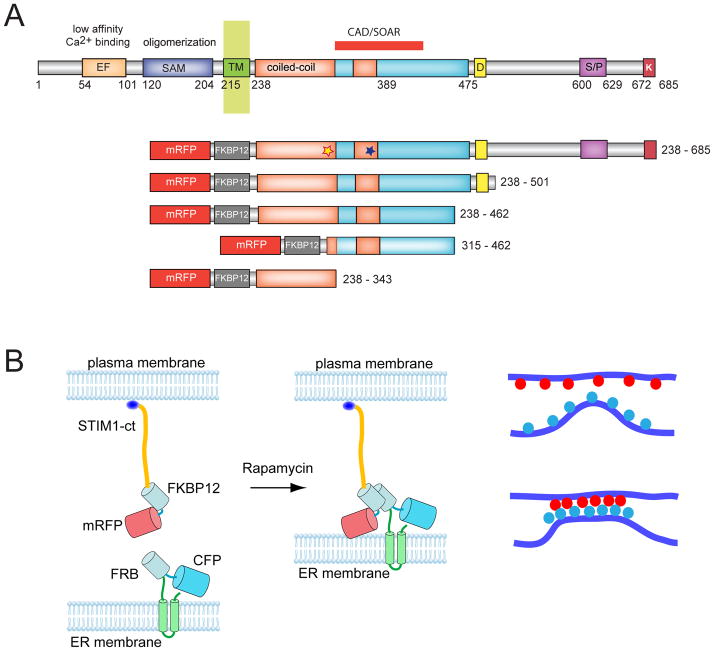
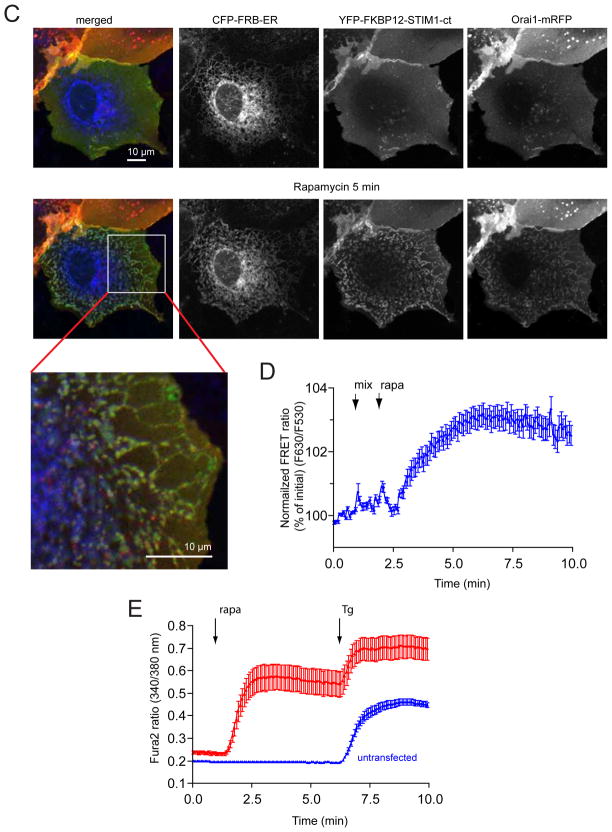
Clustering of the cytosolic domain of STIM1 activates Orai1 channels. (A) Schematics of STIM1 structure and the constructs used in the present study. The numbering corresponds to the human STIM1 protein. EF, Ca2+ binding EF-hand motif; SAM, sterile alpha motif; CAD/SOAR minimal Orai1 activation domain; D, acidic region; S/P, proline-, serine/threonine-rich segment, K, polybasic domain (after(39). The blue and yellow asterisks indicate the positions of the basic and acidic regions, respectively, identified in the present study. (B) Schematics of clustering by rapamycin-induced heterodimerization of the FKBP12 fused cytosolic STIM1 (FK-STIM1-ct) and the FRB targeted to the ER surface. The blue oval represents the C-terminal polybasic domain of STIM1 that is important for the PM localization of the protein (C) Localization of the indicated proteins expressed in COS-7 cells before (top row) and 5 min after (bottom row) rapamycin addition. Confocal images were taken in live cells 1 day after transfection. The area in the white box is shown enlarged. Note the significant membrane localization of the cytoplasmic STIM1 segment and its co-clustering with the Orai1 at the ER-PM contact zones after addition of rapamycin (100 nM). (D) Rapamycin-induced clustering increases FRET between YFP- and mRFP-tagged recruitable STIM1-ct indicating that clustering forces them to get within FRET distance. (E) Cytosolic Ca2+ increases evoked by rapamycin-induced clustering of FK-STIM1-ct. COS-7 cells were transfected with the ER-targeted CFP-FRB, the mRFP-FKBP12-STIM1-ct and untagged Orai1. Cytosolic Ca2+ changes were followed by Fura2. Blue represent cells with no visible transfections. Means ± S.E.M. are shown (n=51 and 58 cells for red and blue, respectively, form two separate experiments).