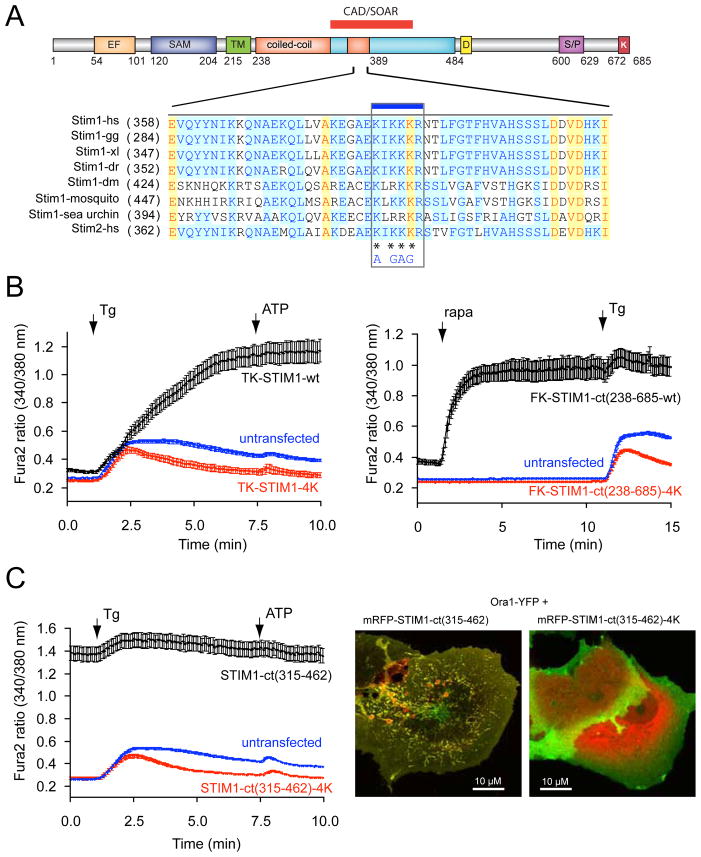
Figure 4.
A polybasic segment within the CAD/SOAR domain is responsible for Orai1 activation. (A) sequence alignment between various STIM1 orthologues and STIM2 within the CAD/SOAR domain (gg, Gallus gallus; xl, Xenopus laevis; dr, danio rero; dm, Drosophila melanogaster. The mutated Lys residues are labeled with an asterisk. (B) The 4K mutation renders full-length STIM1 proteins inactive. COS-7 cells were transfected with the thimidine-kinase (TK) driven STIM1 (black) or its 4K mutant (red) together with untagged Orai1 and their cytoplasmic Ca2+ monitored by Fura2. Thapsigargin (Tg, 200 nM) was added to empty the Ca2+ stores. Means ± S.E.M. are shown (n = 135, 132 and 160 cells for black, red and blue traces, respectively, obtained in 4–5 separate experiments). (C) The same 4K mutation makes the STIM1-ct fragment inactive not responding to clustering with Ca2+ increase (red). Means ± S.E.M. are shown (n = 170, 189 and 239 cells for black, red and blue traces, respectively obtained in 6–7 separate experiments). (D) The 4K(AGAG) mutation inactivates the otherwise constitutively active STIM1-ct(315–462) segment. Means ± S.E.M. are shown (n = 188, 163 and 244 cells for black, red and blue traces, respectively, obtained in 5–7 separate experiments). (E) The constitutively active STIM1(315–462) piece causes clustering and co-localization with Orai1 in COS-7 cells. However, the 4K mutation prevents clustering of Orai1 and the STIM1 construct is largely remains in the cytosol. Confocal images of live cells are shown one day after transfection.