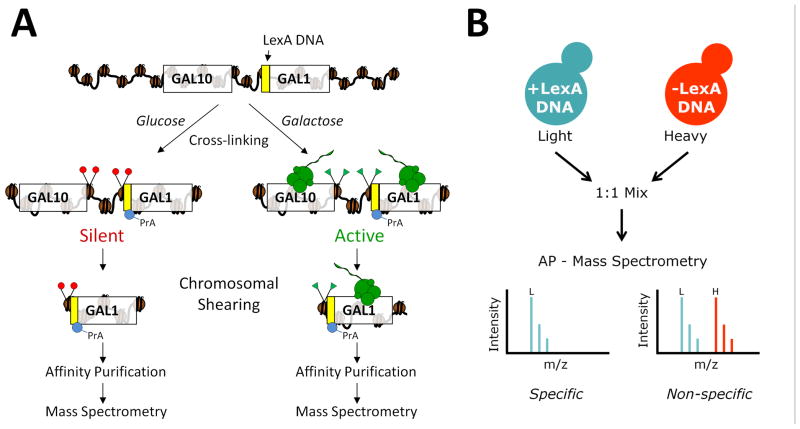
Figure 1. Chromatin affinity purification with mass spectrometry (ChAP-MS).
(A) The ChAP-MS approach provides for the specific enrichment of a given chromosome section and identification of specifically associated proteins and posttranslational modifications. A LexA DNA affinity handle was engineered just upstream of the GAL1 start codon in S. cerevisiae. Strains containing the LexA DNA binding site and a plasmid expressing LexA-PrA protein affinity handle were cultured in glucose or galactose to provide transcriptional repression or activation, respectively, and subjected to in vivo chemical cross-linking to trap protein interactions. Following shearing of the chromatin to ~1,000 bp, LexA-PrA was affinity purified on IgG-coated Dynabeads and co-enriched proteins/posttranslational modifications were identified by high resolution mass spectrometry. (B) To control for non-specifically enriched proteins, a strain lacking the LexA DNA binding site, but containing the LexA-PrA plasmid, was cultured isotopically heavy (13C615N2-lysine) in glucose or galactose and mixed equally with the corresponding isotopically light culture containing the LexA DNA binding site prior to cell lysis. Following affinity purification (AP) and mass spectrometric analysis, non-specifically enriched proteins were identified as a 1:1 ratio of light to heavy lysine-containing peptides, while proteins specifically enriched with the chromosome section were identified with a higher level of isotopically light lysine-containing peptides.