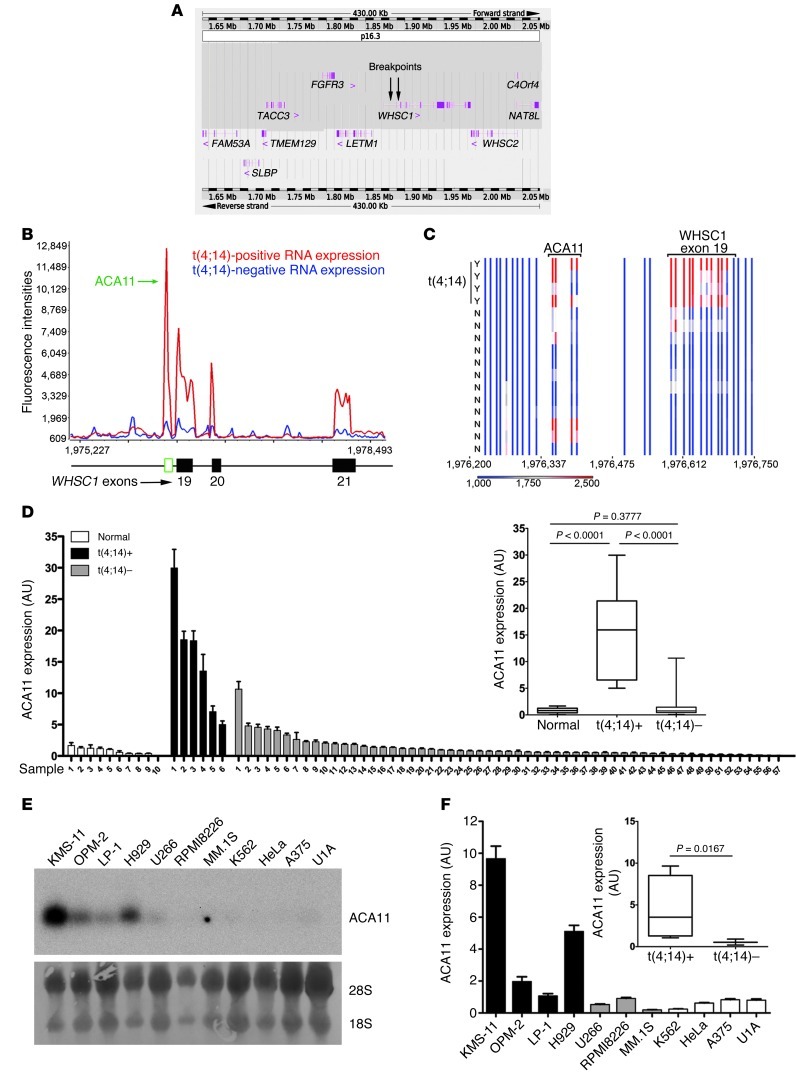
Figure 1. ACA11 is activated by t(4;14) in MM.
(A) Schematic diagram of the chromosome 4p16.3 tiling oligonucleotide microarray, spanning 430 Kb across the t(4;14) breakpoint region (breakpoints are indicated by arrows). Annotated genes are indicated with exon structures. (B) Tiling microarray expression data of representative t(4;14)-positive and -negative patients with MM. (C) Heat map of t(4;14)-positive (Y; n = 4) and -negative (N; n = 12) patient samples. (B and C) Numbers indicate base pairs on chromosome 4 (human genome assembly GRCh37). (D) Real-time PCR of ACA11 in samples from patients with MM. The inset shows that ACA11 is significantly associated with t(4;14) status. Normal, normal plasma cells (n = 10); t(4;14)+, t(4;14)-positive patient samples (n = 6); t(4;14)–, t(4;14)-negative patient samples (n = 57). (E) Northern blot of ACA11 expression in human MM cell lines. t(4;14)-positive cell lines included KMS-11, OPM-2, LP-1, and H929 cells; t(4;14)-negative cell lines included U266, RPMI8226, and MM.1S cells; and non-MM cell lines included K562, HeLa, A375, and U1A cells. Loading controls were 28S and 18S rRNA. (F) ACA11 expression by real-time PCR is significantly associated with t(4;14) in MM cell lines. qRT-PCR data in D and F were normalized to the mean of 3 reference genes (GAPDH, UBC, YWHAZ). Data are presented as mean ± SD, with a box-whisker plot of minimum-to-maximum values (inset) (n = 3). The line within the box is the median value. The box represents the first and third quartiles. The whiskers show the largest and smallest events within at least 1.5 times the size of the box from the nearest edge.