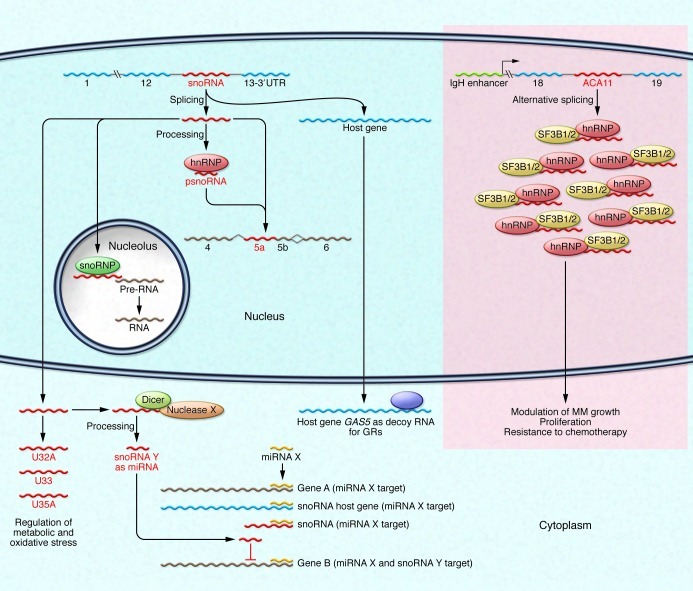
Figure 1. snoRNAs are often localized in introns of RP genes.
snoRNAs are generally involved in rRNA modifications (methylation and pseudouridylation), but orphan snoRNAs are encoded in genes not enriched in specific functional categories. ACA11 orphan snoRNA is encoded within intron 18–19 of the WHSC1 gene and is overexpressed in t(4;14)-positive patients with MM. ACA11 modulates the growth of MM cells, resistance to chemotherapy, and oxidative stress. Other orphan snoRNAs (U32A, U33, U35A) localize in the cytoplasm and regulate metabolic and oxidative stress. Interestingly, some snoRNA host genes are lncRNAs and act as decoy RNAs for GRs. Orphan snoRNAs can be involved in splicing or can be processed into smaller snoRNAs (psnoRNAs) and miRNAs. psnoRNAs modulate alternative splicing, whereas snoRNAs with miRNA-like function downmodulate specific mRNAs at the posttranscriptional level. snoRNAs with miRNA-like function can increase the number of miRNAs able to silence genes containing that specific miRNA binding site. In parallel, overexpression of snoRNAs and snoRNAs host genes with miRNA responsive elements can oppose miRNA-dependent silencing by increasing the amount of target RNAs. Thus, orphan snoRNAs and their host genes can act as novel players in the network of ceRNAs in the cell. Delocalization, overexpression, deletions, or point mutations of specific snoRNAs can profoundly deregulate cellular homeostasis and result in transformation via various mechanisms, including cellular stress and posttranscriptional gene silencing. The mechanism by which ACA11 and U32A, U33, and U35A influence metabolic and oxidative stress is still unknown. snoRNP, small nucleolar ribonucleoprotein; SF3B1/2, RNA splicing factors.