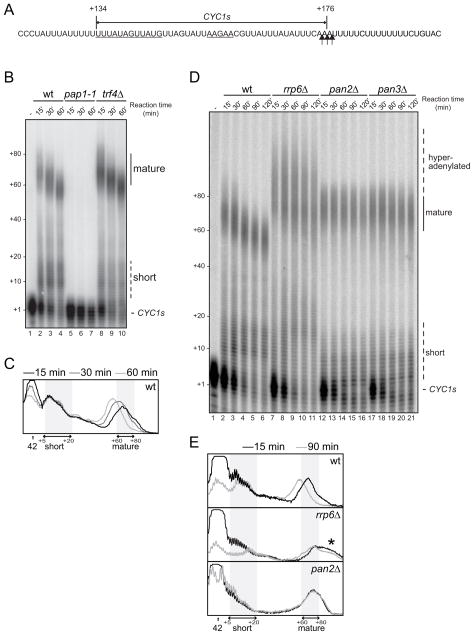
Figure 1. Rrp6p is required for normal pA tail length.
(A) The sequence of the CYC1s substrate used in this study is indicated by a double-arrowed line above the relevant CYC1 3′UTR sequence. Numbering is relative to the CYC1 ORF stop codon. Polyadenylation efficiency- (left) and positioning- (right), elements are underlined. Major cleavage sites are marked by vertical arrows.
(B) CYC1s in vitro polyadenylation time course in wt, pap1-1 and trf4Δ extracts. Positions of precursor (CYC1s), short and mature polyadenylated species are indicated to the right and pA tail lengths estimated from the migration of DNA size markers are shown to the left of the gel.
(C) Phosphorimager lane scans from wt reactions shown in (B) carried out for 15 min (black), 30 min (dark gray) or 60 min (light gray). The position of the precursor (42 nt) is indicated by an arrow. Areas covering short (5–20 nt) and mature (60–80 nt) pA tail regions are marked by double arrows and shaded.
(D) CYC1s in vitro polyadenylation time course in wt, rrp6Δ, pan2Δ and pan3Δ extracts. Image labeling as in (B). ‘Hyperadenylated’ denotes excess adenylation over the normal mature (wt) length.
(E) Lane scans of 15 min (light gray) and 90 min (dark gray) time points of wt (top), rrp6Δ (middle) and pan2Δ (bottom) reactions from (D). Image labeling as in (C). The region of hyperadenylation is indicated by an asterisk.