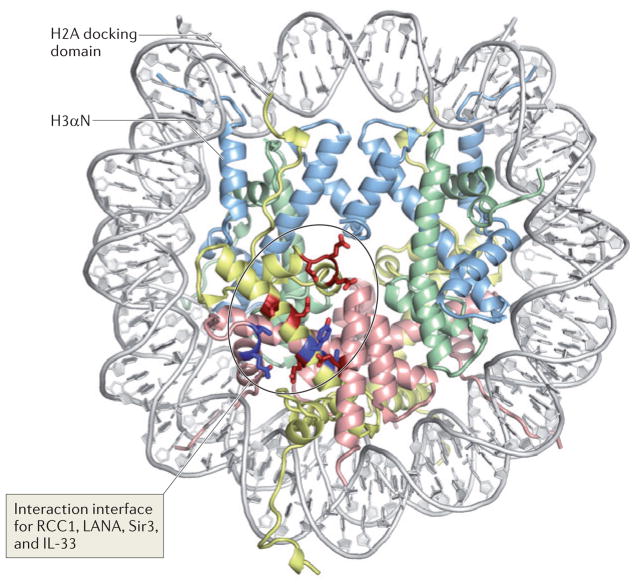
Figure 2. Nucleosome structure and the acidic patch: a common interaction interface for many nucleosome-interacting proteins.
The structure of the nucleosome (Protein Data Bank code 1AOI) is viewed down the superhelical axis of the DNA. Histones H3, H4, H2A and H2B are shown in light blue, green, yellow and red, respectively. The figure indicates the amino-terminal α-helix of H3 (H3αN), which organizes the penultimate 10 bp of the DNA, and the carboxy-terminal end of the H2A docking domain. Acidic residues on H2A and H2B (the ‘acidic patch’) that are involved in the interaction with the H4 tail and with nucleosome-interacting proteins (such as the latency-associated nuclear antigen (LANA) peptide, interleukin-33 (IL-33), regulator of chromosome condensation 1 (RCC1), silent information regulator 3 (Sir3) and high mobility group nucleosome-binding domain-containing protein 2 (HMGN2)) are indicated in bright red; additional residues that are implicated in the interaction interfaces with the proteins listed above are shown in dark blue. Note that the number of total histone residues implicated in all of these protein–protein interfaces is relatively small, and that all cluster in a contained region on the surface of the histone octamer. In the absence of these factors, the interaction of the H4 tail from a neighbouring particle with the acidic patch mediates nucleosome–nucleosome interactions, thereby promoting chromatin folding. By using similar interactions with acidic patch on the nucleosome, the proteins listed above may compete with the H4 tail and modulate chromatin structure.