Abstract
Claspin is a key mediator of the ATR-Chk1 checkpoint pathway. In response to DNA damage, Claspin interacts with Rad17 and Chk1 in a phosphorylation-dependent manner, enabling ATR to phosphorylate Chk1 efficiently. Claspin also interacts with Rad9, but how they interact and whether the interaction is functional remains unknown. Unexpectedly, our analysis of two splicing isoforms of Claspin provided an answer to these questions. The Claspin1339 isoform contains an evolutionarily conserved C terminus, but the Claspin1332 isoform does not. Although the transcripts encoding both Claspin isoforms coexist in HCT116 cells, Claspin1339 is the predominant form responsible for Chk1 activation. When expressed in cells depleted of endogenous Claspin, both Claspin1339 and Claspin1332 are able to mediate Chk1 activation. However, the activation of Chk1 is delayed in Claspin1332-expressing cells compared with Claspin1339-expressing cells. Furthermore, only Claspin1339 but not Claspin1332 interacts with Rad9 efficiently. Together, these results suggest that the conserved C terminus of Claspin interacts with Rad9 and ensures timely activation of the ATR-Chk1 pathway.
Keywords: Chk1, Claspin, Rad9, isoforms, splicing
Introduction
The DNA damage checkpoint is a barrier against genomic instability and tumorigenesis.1,2 In response to DNA damage, the checkpoint is activated to arrest the cell cycle, promote DNA repair or trigger cell death. The ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) kinases are key regulators of the checkpoint.3,4 Whereas ATM primarily responds to double-stranded DNA breaks (DSBs), ATR is activated by a wide spectrum of DNA damage and DNA replication stress. ATR and its functional partner ATRIP are recruited to RPA-coated single-stranded DNA (ssDNA) at sites of DNA damage or stalled replication forks,5 and its kinase activity is stimulated by TopBP1.6 Once activated, ATR phosphorylates and activates the effector kinase Chk1 through a process that depends on the mediator protein Claspin.7,8 The ATR-Claspin-Chk1 pathway plays a critical role in protecting DNA replication forks against DNA damage.
Claspin is a protein involved in both DNA replication and the response to DNA damage.8-11 When DNA damage interferes with DNA replication, Claspin interacts with Rad17, a DNA damage sensor required for ATR activation.12,13 Furthermore, Claspin is phosphorylated by CK1 in an ATR-dependent manner, increasing the affinity of Claspin to Chk1.14-16 In vitro, Claspin stimulates the phosphorylation of Chk1 by ATR.17,18 How Claspin regulates Chk1 activation in human cells is not yet fully understood, and it remains an important question for the research of DNA damage response.
The human CLSPN gene encoding Claspin is located on chromosome 1p34.2.7 Several isoforms of the CLSPN transcript are included in the GeneBank. Among these, the originally reported CLSPN transcript (AF297866.1) encodes a protein of 1332 amino acids (referred to as Claspin1332),7 whereas the subsequently deposited CLSPN transcript 1 (NM_022111.3) encodes a protein of 1339 amino acids (referred to as Claspin1339). Claspin1332 and Claspin1339 are only distinct in their extreme C termini (see below). The C terminus of Claspin1339 but not Claspin1332 is evolutionarily conserved. Both Claspin1332 and Claspin1339 have been widely used in functional analyses by previous studies9,10,18-24. However, which of these Claspin isoforms is predominant in cells and whether they are equally functional is not clear.
Here, we show that the CLSPN transcripts encoding the two distinct C termini of Claspin coexist in multiple human cancer cell lines. In the colon cancer cell line HCT116, the predominant form of Claspin contains the conserved C terminus. When expressed in cells depleted of endogenous Claspin, both Claspin1339 and Claspin1332 are able to mediate Chk1 activation in response to UV radiation. However, Chk1 activation in cells expressing Claspin1332 is delayed compared with cells expressing Claspin1339. In addition, unlike Claspin1339, Claspin1332 is unable to efficiently interact with Rad9, a component of the Rad9-Rad1-Hus1 (9-1-1) complex associated with Rad17. Together, these results suggest that the conserved C terminus of Claspin is a regulatory domain that engages 9-1-1 and promotes the early phase of Chk1 activation.
Results
The CLSPN transcripts encoding distinct C termini coexist in human cells
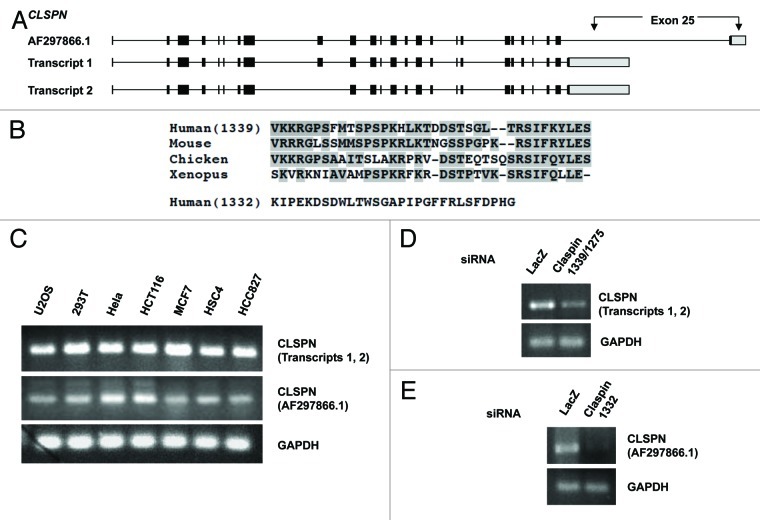
The human CLSPN gene was identified based on the homology between its annotated protein product and Xenpous Claspin protein.7 The originally reported CLSPN transcript (AF297866.1) contains 25 exons, and it encodes a protein of 1332 amino acids (Claspin1332). Two additional CLSPN transcript isoforms were subsequently deposited to the GeneBank. The CLSPN transcript 1 (NM_022111.3) contains 25 exons, whereas the CLSPN transcript 2 (NM_001190481.1) contains only 24 exons. The exon 9 of CLSPN transcript 1 is absent in the transcript 2 (Fig. 1A). The CLSPN transcripts 1 and 2 encode two Claspin isoforms of 1339 (Claspin1339) and 1275 amino acids (Claspin1275), respectively. The CLSPN transcript AF297866.1 and transcript 1 contain different exons 25 (Fig. 1A). As a result, the last 36 amino acids of Claspin1339 and the last 29 amino acids of Claspin1332 are distinct (Fig. 1B). The extreme C terminus of Claspin1339 but not Claspin1332 is conserved in vertebrates (Fig. 1B), suggesting that Claspin1332 is either a recently emerged variant or a product in certain cancer cells.
Figure 1. The Claspin transcript isoforms encoding distinct C termini. (A) A schematic representation of the CLSPN transcript isoforms. (B) Alignment of the C terminus of human Claspin1339 with the corresponding regions in Claspin homologs from other species. The conserved amino acids are shaded. The C terminus of human Claspin1332 is also shown. (C) The CLSPN transcripts encoding the two distinct C termini were detected in a panel of human cancer cell lines by RT-PCR using specific primers. RT-PRC of GAPDH served as the control for mRNA inputs. (D and E) HCT116 cells were transfected with si1339/1275 and si1332, and mRNA was prepared and analyzed by RT-PCR as in (C).
To distinguish the CLSPN transcripts encoding the two distinct C termini, we designed two primer sets to specifically amplify the two alternative exons 25. The primer set 1 detects both the CLSPN transcripts 1 and 2, whereas the primer set 2 specifically detects the CLSPN transcript AF297866.1. RT-PCR was performed using the two primer sets and mRNA derived from a panel of human cancer cell lines (Fig. 1C). The CLSPN transcripts encoding the two distinct C termini were detected in all cell lines, suggesting that they are present broadly and not mutually exclusive. To confirm the specificity of RT-PCR, we designed two siRNAs (si1339/1275 and si1332) that specifically target the two alternative exons 25 in the CLSPN transcripts (Fig. 1D and E). The two siRNAs effectively reduced the levels of their respective target mRNAs in HCT116 cells.
Together, these results suggest that the CLSPN transcripts encoding the two distinct C termini coexist in human cells. Furthermore, given that the cancer cell lines that we analyzed have different origins, the presence of the CLSPN transcript AF297866.1 in all these cell lines suggests that Claspin1332 is unlikely associated with a specific oncogenic event. However, we noted that the relative abundance of the CLSPN transcripts varied among the cell lines. Whether the ratio of Claspin isoforms is functionally relevant remains to be investigated.
Both Claspin1339 and Claspin1332 are functional for Chk1 activation
Using the siRNAs that specifically target the Claspin isoforms with distinct C termini, we next investigated the relative abundance and functional contribution of these proteins. Transfection of HCT116 cells with a pan-isoform Claspin siRNA (siPAN) that targets all three isoforms significantly reduced the levels of Claspin protein (Fig. 2A). Consequently, the UV-induced Chk1 phosphorylation was diminished in siPAN-treated cells (Fig. 2A). Similar to siPAN, si1339/1275, which targets the conserved C terminus, also significantly reduced the overall levels of Claspin protein and UV-induced Chk1 phosphorylation (Fig. 2A). In marked contrast, si1332, which targets the unconserved C terminus, did not reduce the levels of Claspin protein, nor did it reduce the UV-induced Chk1 phosphorylation (Fig. 2A). These results suggest that Claspin1339 and/or Claspin1275, but not Claspin1332, are predominant in HCT116 cells and primarily responsible for UV-induced Chk1 activation. As shown in Figure 2B, the electrophoretic migration of endogenous Claspin was identical to that of Flag-tagged Claspin1339, suggesting that Claspin1339 is the predominant isoform.
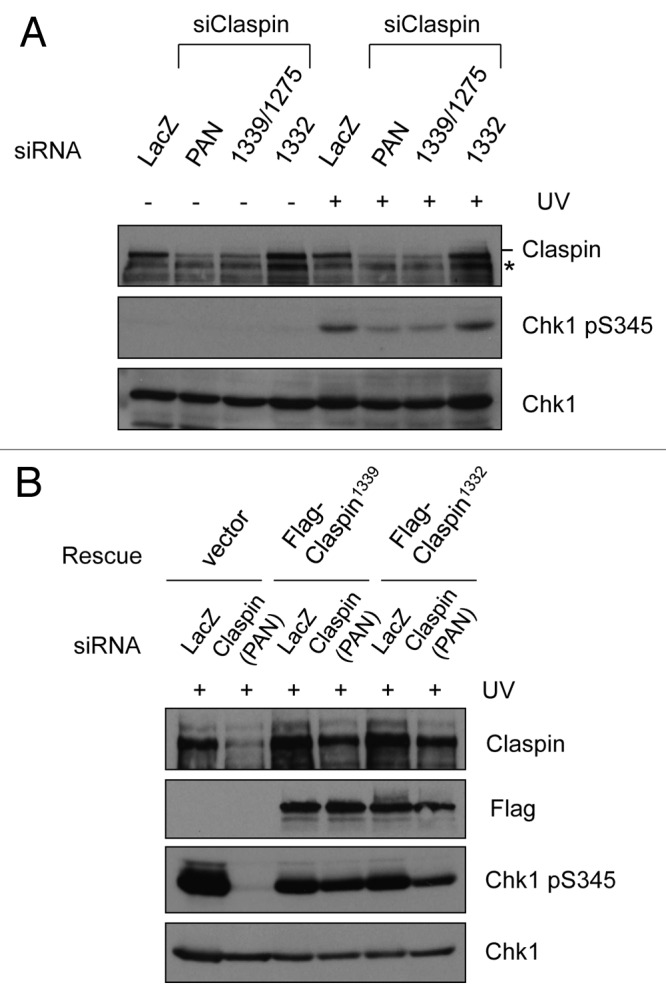
Figure 2. Both Claspin1339 and Claspin1332 are functional for Chk1 activation. (A) HCT116 cells were transfected with siPAN, si1339/1275 or si1332, and irradiated with 15 J/m2 UV after 48 h. The phosphorylation of Chk1 and the levels of the indicated proteins were analyzed by western blot 1 h after UV irradiation. *, a protein that cross-reacted with the Claspin antibody. (B) HCT116 cells were first transfected with siLacZ or siPAN, and then transfected again with empty vector, or plasmids expressing Flag-Claspin1339 or Flag-Claspin1332. Cells were irradiated with 15 J/m2 UV, and the levels of phosphorylated Chk1 and the indicated proteins were analyzed by western blot 1 h after UV damage.
Since both Claspin1332 and Claspin1339 have been used in functional analyses in previous studies, we asked if these Claspin isoforms were equally functional. We generated plasmids that express Flag-tagged Claspin1332 or Claspin1339. The coding sequences of Claspin in these plasmids were modified to render them resistant to siPAN. When Flag-Claspin1332 and Flag-Claspin1339 were individually expressed in siPAN-treated cells, they both suppressed the defect in Chk1 phosphorylation 1 h after UV irradiation (Fig. 2B). In contrast, Chk1 phosphorylation remained defective in the siPAN-treated cells that were subsequently transfected with empty vector. These results suggest that, although the abundances of Claspin1332 and Claspin1339 are different in cells, they are both functional in mediating Chk1 activation.
Cells expressing Claspin1332 activate Chk1 with a delay
In the experiment where we compared the abilities of Claspin1332 and Claspin1339 to mediate Chk1 activation (Fig. 2B), we noticed that UV-induced Chk1 phosphorylation was slightly more efficient in Claspin1339-expressing cells than in Claspin1332-expressing cells. To compare the functions of these Claspin isoforms more carefully, we performed the same experiment in Figure 2B and monitored Chk1 phosphorylation at 0, 0.5, 1, 2, 4 and 8 h after UV irradiation. In Claspin1339-expressing cells, Chk1 became maximally phosphorylated 1 h after UV irradiation (Fig. 3A). However, in Claspin1332-expressing cells, Chk1 phosphorylation did not reach its plateau until 4 h after UV irradiation (Fig. 3B). Thus, although Claspin1332 is capable of mediating Chk1 activation, there is a delay in the timing of Chk1 activation when Claspin1332 is the only form of Claspin in cells.
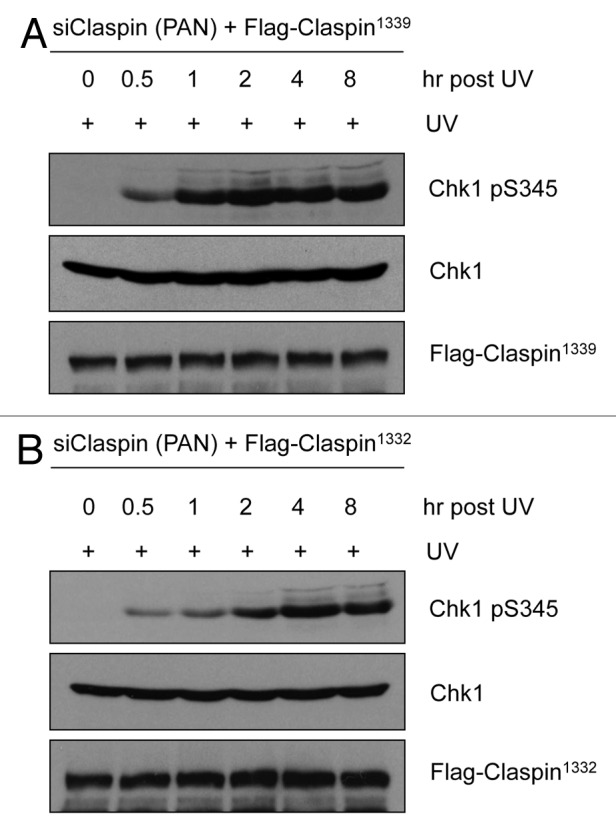
Figure 3. Chk1 phosphorylation is delayed in Claspin1332-expressing cells. (A and B) HCT116 cells were sequentially transfected with siPAN and plasmids expressing Flag-Claspin1339 or Flag-Claspin1332 vector. Cells were irradiated with 15 J/m2 UV, and cells extracts were prepared and analyzed by western blot at the indicated time points.
Only Claspin1339 but not Claspin1332 interacts with Rad9
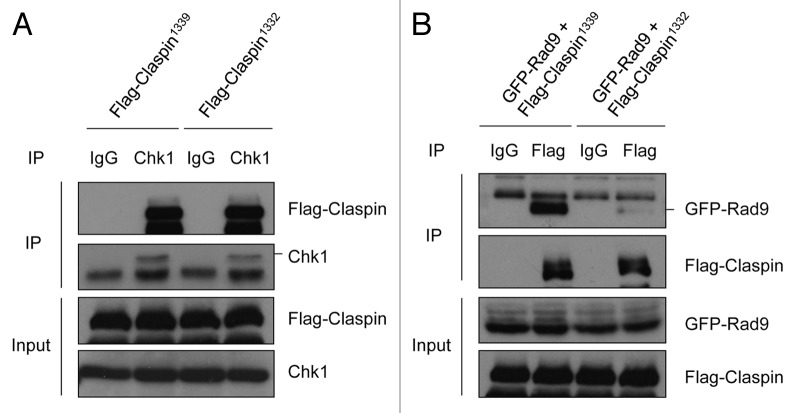
Given the delay of Chk1 activation in Claspin1332-expressing cells, we postulated that Claspin1332 might be defective in its interaction with another checkpoint protein. Claspin is known to interact with Chk1 in a DNA damaged-induced manner.7 To compare the abilities of Claspin1332 and Claspin1339 to interact with Chk1, we transiently expressed Flag-Claspin1332 or Flag-Claspin1339 in HEK293T cells and then irradiated the cells with UV (Fig. 4A). Immunoprecipitation of endogenous Chk1 from cell extracts captured both Flag-Claspin1332 and Flag-Claspin1339 with similar efficiency (Fig. 4A). In contrast, neither Claspin isoform was detected in the control immunoprecipitation with IgG. This experiment rules out the possibility that the two Claspin isoforms have different binding affinities to Chk1 after UV damage.
Figure 4. Claspin1339 but not Claspin1332 interacts with Rad9 efficiently. (A) HEK293T cells transiently expressing Flag-Claspin1339 or Flag-Claspin1332 were irradiated with 15 J/m2 UV and lysed after 1 h. Cell extracts were subjected to immunoprecipitation with Chk1 antibodies. The Chk1 and Flag-tagged Claspin in input and precipitates were analyzed by western blot. (B) HEK293T cells co-expressing GFP-Rad9 and Flag-Claspin1339 or Flag-Claspin1332 were irradiated with UV and analyzed by immunoprecipitation using Flag antibodies. The levels of GFP-Rad9 and Flag-tagged Claspin in input and precipitates were analyzed by western blot.
In addition to Chk1, Claspin is also known to interact with Rad9,25 a component of the 9-1-1 complex that is recruited to damaged DNA by the Rad17-RFC complex. To compare the abilities of Claspin1132 and Claspin1139 to interact with Rad9, we transiently expressed GFP-Rad9 with either Flag-Clapsin1339 or Flag-Claspin1332 in HEK293T cells, irradiated the cells with UV and then immunoprecipitated Flag-tagged Claspin from cell extracts (Fig. 4B). Only Flag-Claspin1339 but not Flag-Claspin1332 efficiently captured GFP-Rad9 from extracts, suggesting that the conserved C terminus of Claspin is important for the interaction with Rad9. Furthermore, these results suggest that the delay of Chk1 activation in Claspin1332-expressing cells may be attributed to the defective Claspin1332-Rad9 interaction.
Discussion
Claspin is a dual functional protein that is involved in both DNA replication and the response to DNA damage. Even in the absence of extrinsic DNA damage, Claspin plays an important role in the progression and stabilization of DNA replication forks, suggesting that Claspin may be a component of the DNA replication machinery.8,11,26,27 When DNA replication forks encounter DNA damage, increased amounts of ssDNA are generated, leading to recruitment of RPA and the Rad17-RFC complex to stalled replication forks.12,28,29 The Rad17-RFC complex recognizes the junctions between ssDNA and dsDNA (double-stranded DNA) and loads the 9-1-1 complexes onto damaged DNA.28,30 Independently of Rad17, RPA coated ssDNA directly recruits the ATR-ATRIP complex and promotes ATR autophosphorylation at Thr 1989.5,31 Together, the 9-1-1 complex and phosphorylated ATR on damaged DNA bring TopBP1 to the protein complex, which stimulates the kinase activity of ATR-ATRIP and facilitates ATR to recognize its substrates.
How does Claspin contribute to checkpoint activation? Claspin is known to interact with phosphorylated Rad17, a substrate of ATR.13 This Rad17-Claspin interaction may be established at stalled replication forks after the initial activation of ATR, promoting the subsequent phosphorylation of Claspin by ATR. In an ATR-dependent manner, Claspin is phosphorylated by CK1 at its C terminus, creating a binding site for Chk1.16 The phosphorylation-mediated interaction between Claspin and Chk1 may transiently recruit Chk1 to stalled replication forks, stimulating the phosphorylation of Chk1 by ATR and Chk1 autophosphorylation.17,18 In this study, we found that Claspin engages Rad9 through the conserved C terminus. Although this interaction is not absolutely required for Chk1 activation, it is important for efficient Chk1 phosphorylation during the early phase of ATR response. In Xenopus egg extracts, Rad17 and 9-1-1 remain associated after DNA damage.32 It is possible that the interaction between Claspin and Rad9 enables Claspin to interact with Rad17 more efficiently, thereby accelerating Chk1 recruitment and subsequent activation. Our findings suggest that the protein-protein interactions within the checkpoint-signaling complex control not only the magnitude but also the timing of checkpoint response. To our knowledge, the conserved C terminus of Claspin is the first regulatory domain of a checkpoint protein that has a clear role in the temporal control of DNA damage signaling.
Our results also raise the interesting possibility that alternative splicing of the CLSPN transcript may influence the kinetics of ATR response. Given that Claspin1332 mediates Chk1 activation with a delay, its presence in replication forks may exert dominant negative effects on certain early events during activation of the ATR-Chk1 pathway. The rapid response of ATR pathway may be important for stabilizing stalled replication forks and promoting specific DNA damage repair or tolerance events. The inability of cells to activate the ATR pathway promptly may compromise genomic stability. The relative abundance of different CLSPN transcripts appears to vary among different cancer cell lines (Fig. 1C). Due to their difference in 3′ UTR, the CLSPN transcripts that contain distinct exons 25 may be subjected to differential regulation by miRNAs or other post-transcriptional regulatory mechanisms. It would be interesting to further investigate whether an altered ratio of the Clapsin1339 and Claspin1332 isoforms is associated with and contributes to tumorigenesis.
Materials and Methods
Cell lines and cell culture
HeLa, HEK293T, U2OS, MCF7, HSC4 and HCC827 cells were maintained in DMEM medium (Invitrogen Corporation) supplemented with 10% fetal bovine serum. HCT116 cells were maintained in McCOY’s 5A medium (Invitrogen) supplemented with 10% fetal bovine serum.
siRNAs and plasmids
siRNAs were transfected with Oligofectamine and plasmids were transfected with Lipofectamine following the manufacturer’s instructions (Invitrogen). The siRNAs targeting LacZ, Claspin (PAN), Claspin (1339/1275) and Claspin (1332) were synthesized by Invitrogen. The siRNA sequences were as follows: LacZ: AACGUACGCGGAAUACUUCGA; Claspin (PAN): AAGGAAAGAAAGGCAGCCAGA; Claspin (1339/1275): GGCCTTGGGTTAGGT TTCACTTCCT; Claspin (1332/1275): CAAAGAGAGGCGGAGAAGGGCTATT.
To express Flag-tagged Claspin1339 in human cells, the Claspin coding sequence was PCR amplified from the I.M.A.G.E. clone 7961377 and cloned into the pFlag-CMV2 vector. Flag-Claspin1332 was generated by cloning the Claspin coding sequence from S-Flag-Claspin (a gift of Dr. Junjie Chen) into the pFlag-CMV2 vector.
Antibodies
Chk1 rabbit polyclonal antibody (sc7898), Chk1 mouse monoclonal antibody (G4, sc8408), GFP rabbit polyclonal antibody (sc8334), normal rabbit IgG (sc-2027) and normal mouse IgG (sc-2025) were from Santa Cruz Biotechnology, Inc. Claspin rabbit polyclonal antibody (A300–267) was from Bethyl Laboratories, Inc. Mouse Flag monoclonal antibody (F1804, M2) was from Sigma-aldrich Corporation. Rabbit pChk1s345 monoclonal antibody was from Cell Signaling Technology, Inc. GFP mouse monoclonal antibody (ab1218) was from Abcam PLC.
Immunoprecipitation
HCT116 cells were lysed in NETN buffer [20 mM TRIS-HCl (pH8.0), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40] containing 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF and a protease inhibitor cocktail (Sigma). Cell extracts were spun at 14,000 rpm for 10 min, and the resulting supernatants were precleared with Protein G/A Sepharose beads. Antibodies to Flag or Chk1 were added to the extracts containing Protein G/A Sepharose beads, and were incubated overnight at 4°C. Subsequently, the Sepharose beads were sedimented and washed four times with NETN buffer. The immunoprecipitates were then subjected to SDS-PAGE and western blot analysis.
RT-PCR
The total mRNA was extracted using the RNeasy Mini kit (Qiagen). After determining the concentration of the mRNA, RT-PCR was performed using the Qiagen OneStep RT-PCR Kit (Qiagen). Primers for the exon 25A are: CTCCTGTCAAGGCTGAGGC and CAAAGCAGTCTCAATGTAG. Primers for the exon 25B are: CTGACCATAACCC AGTGCT and TTGTTCTGCCCAGAATAGCC. Primers for GAPDH are: GCCTCA AGATCATCAGCAATG and CCACGATACCAAAGTTGTCATGG.
Acknowledgment
We thank Drs. William Dunphy, Junjie Chen, Daniel Haber and Jiri Bartek for Claspin plasmids, and members of the Zou lab for helpful discussions. L.Z. is supported by the NIH grant GM076388 and a grant funded by the Federal Share of Proton Program Income. L.Z. is a Jim and Ann Orr MGH Research Scholar.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21041
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–40. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett. 2011;585:1625–39. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 7.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–49. doi: 10.1016/S1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–64. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–52. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chini CC, Wood J, Chen J. Chk1 is required to maintain claspin stability. Oncogene. 2006;25:4165–71. doi: 10.1038/sj.onc.1209447. [DOI] [PubMed] [Google Scholar]
- 11.Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol Biol Cell. 2008;19:2373–8. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–41. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 14.Kumagai A, Dunphy WG. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol. 2003;5:161–5. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- 15.Jeong SY, Kumagai A, Lee J, Dunphy WG. Phosphorylated claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J Biol Chem. 2003;278:46782–8. doi: 10.1074/jbc.M304551200. [DOI] [PubMed] [Google Scholar]
- 16.Meng Z, Capalbo L, Glover DM, Dunphy WG. Role for casein kinase 1 in the phosphorylation of Claspin on critical residues necessary for the activation of Chk1. Mol Biol Cell. 2011;22:2834–47. doi: 10.1091/mbc.E11-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumagai A, Kim SM, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279:49599–608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- 18.Lindsey-Boltz LA, Serçin O, Choi JH, Sancar A. Reconstitution of human claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase. J Biol Chem. 2009;284:33107–14. doi: 10.1074/jbc.M109.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Song YH, Brannigan BW, Wahrer DC, Schiripo TA, Harris PL, et al. Prevalence and functional analysis of sequence variants in the ATR checkpoint mediator Claspin. Mol Cancer Res. 2009;7:1510–6. doi: 10.1158/1541-7786.MCR-09-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serçin O, Kemp MG. Characterization of functional domains in human Claspin. Cell Cycle. 2011;10:1599–606. doi: 10.4161/cc.10.10.15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke CA, Clarke PR. DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J. 2005;388:705–12. doi: 10.1042/BJ20041966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett LN, Clarke PR. Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006;580:4176–81. doi: 10.1016/j.febslet.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 23.Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307–18. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, et al. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–29. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–62. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–40. doi: 10.1016/S1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Focarelli ML, Soza S, Mannini L, Paulis M, Montecucco A, Musio A. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer. 2009;48:1083–90. doi: 10.1002/gcc.20710. [DOI] [PubMed] [Google Scholar]
- 28.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–32. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupardus PJ, Byun T, Yee MC, Hekmat-Nejad M, Cimprich KA. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 2002;16:2327–32. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namiki Y, Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci USA. 2006;103:580–5. doi: 10.1073/pnas.0510223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–35. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]