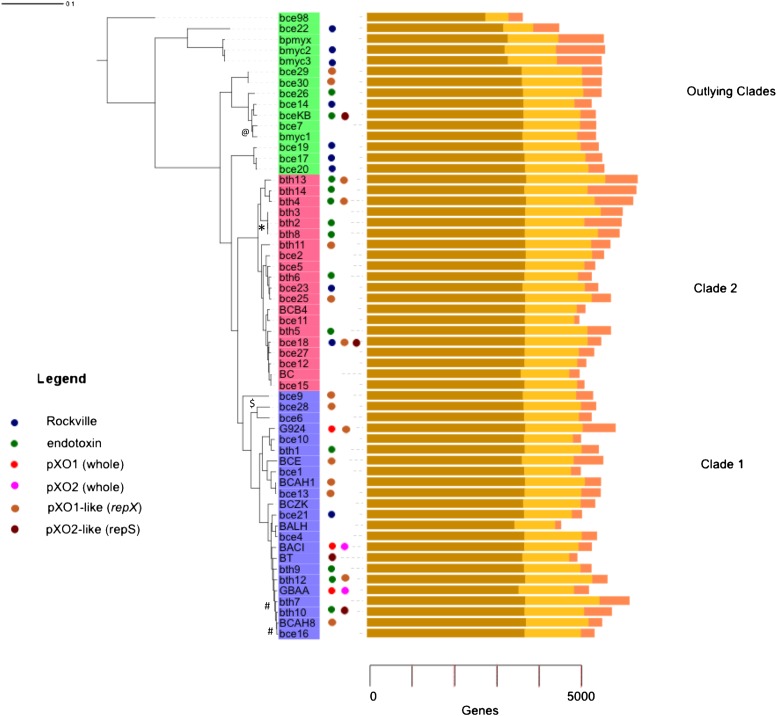
Figure 1.
Whole-genome phylogeny of B. cereus s.l. The phylogenic tree was constructed from a data set of concatenated, conserved protein sequences using the neighbor-joining (NJ) algorithm implemented by PHYLIP (Felsenstein 1989). The tree was rooted using the known outlier B. cereus subsp cytotoxis. The scale measures number of substitutions per residue. Tree topologies computed using maximum likelihood and parsimony estimates are identical with each other and the NJ tree (see Supplemental Data). We performed 1000 bootstrap replications to test the topology of the tree, and all branches were supported by >50% of the trials. All branches with a bootstrap value of >0.9 are labeled with the following codes: 0.8–0.9; “$”; 0.7–0.8, “#”; 0.6–0.7 “@”; 0.5–0.6 “*”. If not labeled, support was >0.9. Labels refer to the genome codes listed in Table 1. Clades 1 and 2 and the outliers are labeled with blue, red, and green strips, respectively. Circles are opposite genomes containing whole pXO1 (red), pXO2 (fuchsia), or δ-endotoxin genes (green) or were isolated on the same day from Rockville, Maryland (blue). We have also indicated genomes likely containing pXO1-like (brown) or pXO2-like (burgundy) plasmids based on the presence of the RepX (Anand et al. 2008) or RepS (Tinsley et al. 2004) proteins, respectively. Each genome has a bar graph showing the proportion of genes belonging to core (brown), character (ocher), and accessory (pink) classes. All unfinished genomes sequenced in this project can be readily identified because they have three-letter lowercase species identifiers. The only other unfinished genome is G9241. Figures 1 and 2 were drawn using online Interactive Tree of Life (iTOL) software (Letunic and Bork 2007).