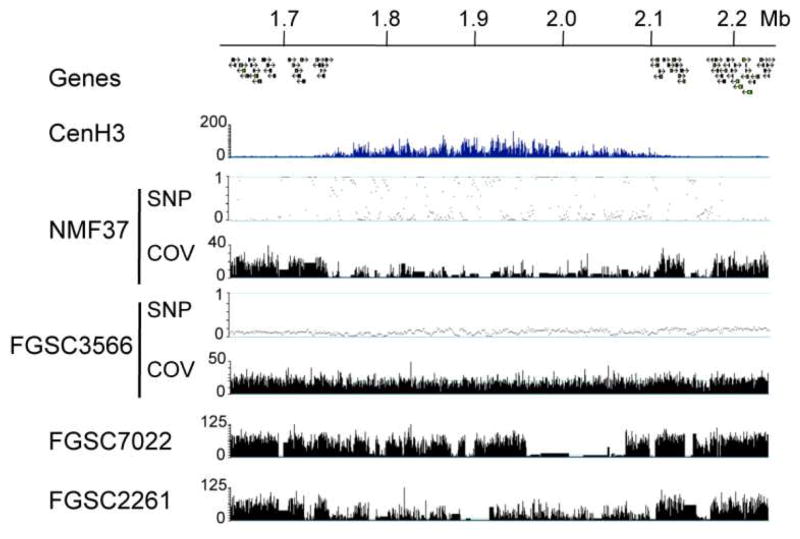
Figure 1. Different Neurospora strains have vastly different DNA sequence at their centromeres.
The centromere region of LG VII is identified by the absence of genes and enrichment of CenH3 by ChIP-seq (“CenH3”). Genomic HTS of strains with diverse genetic backgrounds demonstrates DNA sequence differences in centromere regions. There are regions of missing sequence and high levels of SNPs in the centromere of NMF37, the Mauriceville wild-collected strain. Both FGSC3566 and FGSC7022 have mixed genetic backgrounds, and FGSC2261 was backcrossed into the St. Lawrence lineage two times. Tracks show SNPs per kb (“SNP”) and coverage (“COV”) as read coverage per base (if not specified, tracks show coverage).