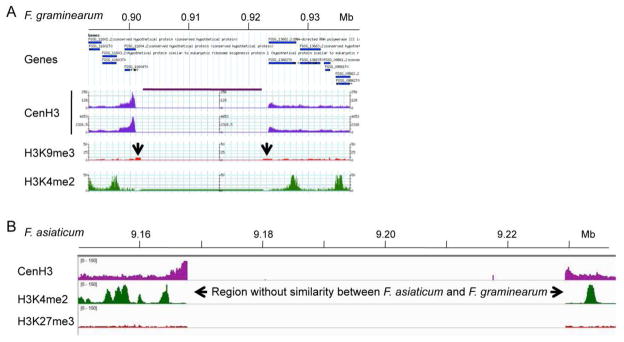
Figure 2. Mapping of Fusarium centromeres by ChIP-seq with CenH3.
A. Region that contains the presumed centromeric DNA on Chromosome 1 of Fusarium graminearum. Genes flanking the centromere are shown in blue, CenH3 ChIP-seq-derived reads are shown in purple (two replicates), enrichment for the silencing H3K9me3 modification is shown in red, and the activating H3K4me2 modification is shown in green. The region under the purple line above the two CenH3 tracks contains only “N”s. The two arrows indicate position of misassembled DNA that cannot be amplified from the genome by PCR. B. Mapping of F. graminearum CenH3 ChIP-seq reads to the F. asiaticum Cen1 region. Only the edges of the centromeric regions are identified. The core region is so divergent that essentially no reads map to this AT- and repeat-rich region. One silencing (H3K27me3, red) and one activating (H3K4me2, green) histone modification are also shown. In both A. and B. CenH3 enrichment was observed at the edges of the presumed centromeric regions (and even overlapping genes, as in A.), likely because shearing by sonication can result in arrays of several nucleosomes.