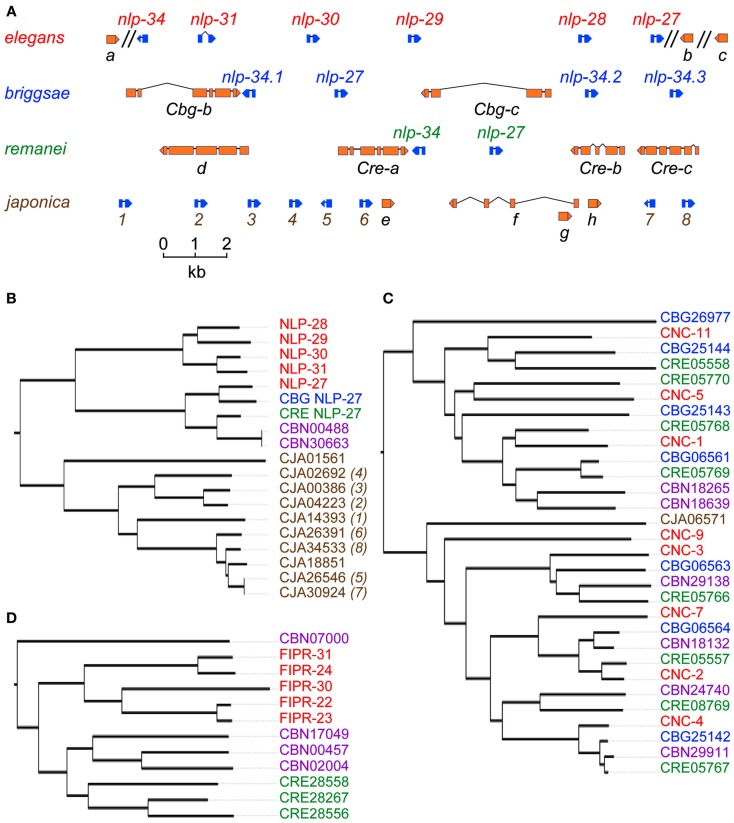
Figure 1.
(A) The nlp-29 cluster in C. elegans together with syntenic regions from C. brigssae (Cbg), C. remanei (Cre), and C. japonica. AMP genes of the nlp class are shown in blue; there has been a marked expansion in C. japonica. The genes labeled “a” and “c” in C. elegans, and their orthologs (Cbg-a, Cbg-c, Cre-a, and Cre-c) are predicted to encode serpentine receptors; “b” in C. elegans is K09D9.9. Its ortholog in C. japonica is ca. 60 kb 3′ of the locus. Immediately 3′ of the gene labeled “e” in C. japonica there is the remnant of a degenerate paralog of K09D9.9. The figure is adapted from data in Wormbase WS230; it does not reproduce the predicted fusion of Cre-b and Cre-c, as this does not appear to be probable. Only the 3′ extremities of a, b, and c are shown. (B–D) Phylogenetic trees for selected members of the NLP (B), CNC (C), and FIPR (D) family peptides, including homologs in C. brenneri (CBN). A distance matrix analysis was performed using the alignment program clustalw2 to generate a guide tree via pairwise and subsequent multiple sequence alignment. This guide tree was then used to produce a true phylogenetic tree that was loaded into the Interactive Tree Of Life v2 software suite (Letunic and Bork, 2011). Partial rooted trees were extracted that corresponding to interesting features within the complete FIP/FIPR/NLP/CNC tree. For the non-elegans peptides, with the exception of CBG NLP-27 and CRE NLP-27, the corresponding gene identifier is given. The numbers in brackets for the C. japonica gene names match the corresponding genes in (A).