Abstract
Severe decline of almond trees has recently been observed in several orchards on the island of Mallorca (Balearic Islands, western Mediterranean Sea). However, the identity of the causal agents has not yet been investigated. Between August 2008 and June 2010, wood samples from branches of almond trees showing internal necroses and brown to black vascular streaking were collected in the Llevant region on the island of Mallorca. Several fungal species were subsequently isolated from the margin between healthy and symptomatic tissue. Five species of Botryosphaeriaceae (namely Botryosphaeria dothidea, Diplodia olivarum, D. seriata, Neofusicoccum australe and N. parvum), Eutypa lata, Phaeoacremonium iranianum and Phomopsis amygdali were identified based on morphology, culture characteristics and DNA sequence comparisons. Neofusicoccum parvum was the dominant species, followed by E. lata, D. olivarum and N. australe. First reports from almond include D. olivarum and Pm. iranianum. Two species are newly described, namely Collophora hispanica sp. nov. and Phaeoacremonium amygdalinum sp. nov.
Keywords: almond dieback, Botryosphaeriaceae, Collophora, Eutypa lata, Phaeoacremonium, Phomopsis amygdali, Prunus dulcis
INTRODUCTION
Almond (Prunus dulcis) is a common crop cultivated in many Mediterranean countries as well as in California (USA), South Africa, and some countries in South America and Australasia. According to the Food and Agriculture Organization (FAO 2010), Spain is the second largest almond producer after California, accounting for 11.9 % of the world’s almond production, which yielded 2.31 million tons in 2009. In Spain, almonds are grown in the south-eastern regions and on the Balearic Islands (western Mediterranean Sea), an important region with 23 432 ha of this crop cultivated in 2007 (4.7 % of the Spanish almond cultivation) (INE 2011).
In summer 2008, severe decline of almond trees was noticed in several orchards on the island of Mallorca (Balearic Islands). Disease symptoms included rapid collapse of branches during mid-summer, chlorosis of leaves, which suddenly wilted and died, as well as bud and shoot dieback. Internal wood symptoms ranged from brown to black vascular streaking, visible in cross sections as spots or circular discolouration of the xylem tissue. Additionally, wedge-shaped necroses were frequently observed. Some affected trees in the orchard died within a few weeks of showing first symptoms.
Different studies have shown that Prunus represents a rich catch-crop for many fungal trunk pathogens. Species of Botryosphaeriaceae (Aplosporella, Botryosphaeria, Diplodia, Dothiorella, Lasiodiplodia, Macrophomina, Neofusicoccum, Spencermartinsia and Sphaeropsis), Calosphaeriaceae (Calosphaeria and Jattaea), Coniochaetaceae (genus Coniochaeta), Diaporthaceae (Phomopsis), Diatrypaceae (Cryptovalsa, Diatrype, Eutypa and Eutypella), Herpotrichiellaceae (Phaeomoniella), Montagnulaceae (Paraconiothyrium), Togniniaceae (Phaeoacremonium) and species of the genus Collophora have been reported on Prunus trees. A list of fungal trunk pathogens isolated from Prunus spp. and their worldwide distribution is shown in Table 1. While several fungal species belonging to a number of genera are well-recognised pathogens of Prunus trees, the causal agent of the severe decline of almond trees on the island of Mallorca is still unknown. Therefore, the objective of this study was to determine the aetiology of trunk diseases associated with wood necroses of almond trees in this region of Spain.
Table 1.
List of fungal trunk pathogens isolated from Prunus spp. and their worldwide distribution.
| Family | Species | Prunus spp. | Country | Reference |
|---|---|---|---|---|
| Botryosphaeriaceae | Aplosporella indica, A. phyllanthina | P. domestica | India | Agarwal et al. (1992) |
| A. pruni | P. armeniaca | Australia | McAlpine (1902) | |
| A. prunicola | P. persica var. nucipersica | South Africa | Damm et al. (2007b) | |
| Botryosphaeria dothidea | P. dulcis | USA | Inderbitzin et al. (2010) | |
| P. persica | Worldwide | Pusey et al. (1995) | ||
| Prunus spp. | USA | Farr et al. (1989) | ||
| Diplodia africana | P. persica | South Africa | Damm et al. (2007a) | |
| D. mutila | P. armeniaca, P. persica | Unknown | Sutton (1980) | |
| P. persica | New Zealand | Laundon (1973) | ||
| D. pinea | P. persica | South Africa | Damm et al. (2007a) | |
| D. rosulata | P. africana | Ethiopia | Gure et al. (2005) | |
| D. roumeguerei var. santonensis | P. laurocerasus | France | Wollenweber & Hochapfel (1941) | |
| D. seriata | P. armeniaca | South Africa | Damm et al. (2007a) | |
| P. dulcis | USA | Inderbitzin et al. (2010) | ||
| P. persica | Worldwide | Pusey et al. (1995) | ||
| P. persica, P. salicina | South Africa | Damm et al. (2007a), Slippers et al. (2007) | ||
| P. persica var. nucipersica | South Africa | Damm et al. (2007a) | ||
| Prunus spp. | USA | Farr et al. (1989) | ||
| Dothiorella sarmentorum | P. armeniaca, Prunus spp. | Europe, North America | Wollenweber (1941) | |
| P. dulcis | USA | Inderbitzin et al. (2010) | ||
| Lasiodiplodia plurivora | P. salicina | South Africa | Damm et al. (2007a) | |
| L. theobromae | P. persica | Worldwide | Pusey et al. (1995) | |
| Prunus spp. | USA | Farr et al. (1989) | ||
| Macrophomina phaseolina | P. dulcis | USA | Inderbitzin et al. (2010) | |
| Neofusicoccum australe | P. armeniaca, P. persica | South Africa | Damm et al. (2007a) | |
| P. dulcis | South Africa | Slippers et al. (2007) | ||
| P. salicina | South Africa | Damm et al. (2007a), Slippers et al. (2007) | ||
| N. mediterraneum, N. nonquaesitum, N. parvum | P. dulcis | USA | Inderbitzin et al. (2010) | |
| N. ribis | Prunus spp. | USA | Farr et al. (1989) | |
| N. vitifusiforme | P. persica, P. salicina | South Africa | Damm et al. (2007a) | |
| Spencermartinsia viticola | P. persica var. nucipersica, P. salicina | South Africa | Damm et al. (2007a) | |
| Sphaeropsis peckii | Prunus spp. | USA | Farr et al. (1989) | |
| Calosphaeriaceae | Calosphaeria africana | P. armeniaca | South Africa | Damm et al. (2008a) |
| Jattaea mookgoponga | P. persica var. nucipersica | South Africa | Damm et al. (2008a) | |
| J. prunicola | P. salicina | South Africa | Damm et al. (2008a) | |
| Coniochaetaceae | Coniochaeta africana | P. salicina | South Africa | Damm et al. (2010) |
| Ca. ambigua | P. armeniaca, P. avium | Moldavia | Popushoi (1971) | |
| Ca. calva | P. avium, P. domestica | Moldavia | Popushoi (1971) | |
| Ca. ligniaria | P. domestica | Moldavia | Popushoi (1971) | |
| Ca. prunicola, Ca. velutina | P. armeniaca, P. salicina | South Africa | Damm et al. (2010) | |
| Diaporthaceae | Phomopsis ambigua | P. salicina | South Africa | Smit et al. (1996) |
| Ps. amygdali | P. dulcis, P. persica | Worldwide | Tuset & Portilla (1989) | |
| Ps. mali, Ps. padina, Ps. parabolica, Ps. perniciosa, Ps. pruni, Ps. prunorum, Ps. stipata, Ps. ribetejana P. dulcis, Prunus spp. | Worldwide | Uecker (1988) | ||
| Ps. theicola, Phomopsis sp. | P. dulcis | Portugal | Diogo et al. (2010) | |
| Diatrypaceae | Cryptovalsa ampelina | P. armeniaca | USA | Trouillas et al. (2010) |
| Prunus spp. | South Africa | Damm et al. (2009) | ||
| Diatrype oregonensis | P. armeniaca | USA | Trouillas et al. (2010) | |
| Eutypa lata | P. avium | Worldwide | Munkvold & Marois (1994) | |
| P. armeniaca | Worldwide | Carter (1957) | ||
| P. dulcis, P. salicina | Worldwide | Carter (1982) | ||
| P. virginiana | Worldwide | English & Davis (1965) | ||
| Eutypella prunastri | P. avium, P. dulcis, P. salicina, P. spinosa | Worldwide | Grove (1935), Ellis & Ellis (1997) | |
| Herpotrichiellaceae | Phaeomoniella dura, Pa. effusa, Pa. prunicola, Pa. zymoides | P. salicina | South Africa | Damm et al. (2010) |
| P. tardicola | P. armeniaca | South Africa | Damm et al. (2010) | |
| Montagnulaceae | Paraconiothyrium africanum | P. persica | South Africa | Damm et al. (2008c) |
| Pc. brasilense | P. persica, P. persica var. nucipersica, P. salicina | South Africa | Damm et al. (2008c) | |
| Pc. variabile | P. persica, P. salicina | South Africa | Damm et al. (2008c) | |
| Togniniaceae | Phaeoacremonium aleophilum | P. armeniaca | South Africa | Mostert et al. (2003, 2006), Damm et al. (2008b) |
| P. pennsylvanica | Canada | Hausner et al. (1992) | ||
| P. persica, P. salicina | South Africa | Damm et al. (2008b) | ||
| Pm. australiense, Pm. fuscum, Pm. griseorubrum, Pm. mortoniae, Pm. prunicolum, Pm. viticola | P. salicina | South Africa | Damm et al. (2008b) | |
| Pm. iranianum, Pm. pallidum, Pm. subulatum, Togninia africana, Tg. griseo-olivacea | P. armeniaca | South Africa | Damm et al. (2008b) | |
| Pm. parasiticum | P. avium | Greece | Rumbos (1986) | |
| P. armeniaca | Tunisia | Hawksworth et al. (1976) | ||
| P. armeniaca | South Africa | Damm et al. (2008b) | ||
| Pm. scolyti | P. armeniaca, P. persica, P. persica var. nucipersica, P. salicina | South Africa | Damm et al. (2008b) | |
| Unknown | Collophora africana, Co. capensis, Co. paarla, | |||
| Co. pallida | P. persica, P. persica var. nucipersica | South Africa | Damm et al. (2008b) | |
| Co. rubra | P. dulcis, P. persica, P. persica var. nucipersica | South Africa | Damm et al. (2008b) |
MATERIALS AND METHODS
Sampling and fungal isolation
A field survey was conducted on almond trees (local cultivars Vivot, Pons, Negre and Totsolet) in the Llevant region on the island of Mallorca (Balearic Islands, western Mediterranean Sea) between August 2008 and June 2010 (Table 2). Wood samples were collected from branches of almond trees with dieback symptoms, including dead shoots, cankers, internal wood necrosis, black vascular streaking or discoloured tissues (Fig. 1).
Table 2.
Names, accession numbers, and collection details of isolates studied.
| Species | Accession no.1 | Location | Collector | GenBank accessions2 | ||||
|---|---|---|---|---|---|---|---|---|
| Botryosphaeria dothidea | Bdo-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN183856 | – | JF503990 | – |
| Collophora hispanica | Col-1, CBS 128566 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | JN808850 | JN808839 | JN808843 |
| Col-3, CBS 128567 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | JN808851 | JN808840 | JN808844 | |
| Col-4, CBS 128568 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | JN808852 | JN808841 | JN808845 | |
| Col-5, CBS 128569 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | JN808853 | JN808842 | JN808846 | |
| Diplodia olivarum | Dol-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN183857 | – | JF693916 | – |
| Dol-2 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN183858 | – | JF693917 | – | |
| Dol-3 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN183859 | – | JF693918 | – | |
| Dol-4 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | ||||||
| Dol-5 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Dol-6 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Dol-7 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Diplodia seriata | Dse-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN183860 | – | JN183850 | – |
| Dse-2 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN183861 | – | JN183851 | – | |
| Dse-3 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN183862 | – | JN183852 | – | |
| Dse-4 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Eutypa lata | Ela-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | – | – | JN183853 | – |
| Ela-2 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | – | – | JN183854 | – | |
| Ela-3 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | – | JN183855 | – | |
| Ela-4 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Ela-5 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Ela-6 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Ela-7 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Neofusicoccum australe | Nau-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN191293 | – | JF421449 | – |
| Nau-2 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN191294 | – | JF421450 | – | |
| Nau-3 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN191295 | – | JF421451 | – | |
| Nau-4 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Nau-5 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Nau-6 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Neofusicoccum parvum | Npa-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2009 | – | JN191296 | – | JF330779 | – |
| Npa-2 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN191297 | – | JF330780 | – | |
| Npa-3 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | JN191298 | – | JF330781 | – | |
| Npa-4 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-5 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-6 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-7 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-8 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-9 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-10 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-11 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-12 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Npa-13 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | ||||||
| Phaeoacremonium | ||||||||
| amygdalinum | Psp-1 | Sant Llorenç, Mallorca, Spain | D. Gramaje, 2009 | JN191301 | JN191305 | – | – | – |
| Psp-2 | Sant Llorenç, Mallorca, Spain | D. Gramaje, 2009 | JN191302 | JN191306 | – | – | – | |
| Psp-3, CBS 128570 | Sant Llorenç, Mallorca, Spain | D. Gramaje, 2009 | JN191303 | JN191307 | – | – | – | |
| Psp-4 | Sant Llorenç, Mallorca, Spain | D. Gramaje, 2009 | JN191304 | JN191308 | – | – | – | |
| Phaeoacremonium | ||||||||
| iranianum | Pir-1 | Sant Llorenç, Mallorca, Spain | D. Gramaje, 2009 | JN191300 | JN191299 | – | – | – |
| Phomopsis amygdali | Pam-1 | Sant Llorenç, Mallorca, Spain | J. Armengol, 2010 | – | – | – | JF693919 | – |
1 CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Diversity Centre, Utrecht, The Netherlands.
2 GenBank accessions correspond to ACT, BT, EF, ITS and GAPDH sequences.
Fig. 1.

Disease symptoms on almond trees on the island of Mallorca associated with fungal trunk pathogens. a, b. Dieback and wilting of branches; c–h. internal symptoms visible when transversal and longitudinal cuts were made in branches used for fungal isolation: black spots and dark brown to black streaking of the xylem tissue (d, h), circular (c, g) or sectorial necrosis (f), and wood discoloration (e).
Wood segments were cut from the affected branches, washed under running tap water, surface-disinfected for 1 min in a 1.5 % sodium hypochlorite solution, and washed twice with sterile distilled water. Small pieces from the margin between healthy and discoloured or decayed wood tissue were plated on malt extract agar (MEA, 2 % malt extract, Oxoid Ltd., England; 1.5 % agar, Difco, USA) supplemented with 0.5 g/L of streptomycin sulphate (MEAS) (Sigma-Aldrich, St. Louis, MO, USA). Plates were incubated at 25 °C in the dark for 14 to 21 d, and all colonies were transferred to 2 % potato-dextrose agar (PDA; Biokar-Diagnostics, Zac de Ther, France). Single spore colonies were derived prior to morphological and molecular identification using the serial dilution method (Dhingra & Sinclair 1995) and stored in 15 % glycerol solution at −80 °C in 1.5 mL cryovials.
Morphological identification and characterisation
Species of Botryosphaeriaceae were identified based on colony and conidial morphology as described by Phillips (2006). In order to enhance sporulation, cultures were amended with sterilised pine needles on 2 % water agar (WA; Biokar-Diagnostics) and incubated at 25 °C under near UV light with a 12 h photoperiod (Philips TDL18W/33) (Slippers et al. 2004). Isolates were examined weekly for formation of pycnidia and conidia in order to record their morphology (size, shape, colour, presence or absence of septa and cell wall structure).
Since it is difficult to distinguish species or even genera within the Diatrypaceae based on morphological characters of their Libertella anamorph (Glawe & Rogers 1984), the morphological identification of Eutypa lata was only tentative using characters such as conidial size and shape, and colony characters on PDA.
Morphological characters used to distinguish Phaeoacremonium species included conidiophore morphology, phialide type and shape, size of hyphal warts and conidial size and shape. Colony characters and pigment production on MEA, PDA and oatmeal agar (OA; 60 g oatmeal; 12.5 g agar; Difco, France) (Crous et al. 2009) incubated at 25 °C were noted after 8 and 16 d.
Collophora species were characterised based on the presence of conidiomata, microcyclic conidiation or endoconidia additional to conidia formed on hyphae, as well as size and shape of conidia and conidiophores (Damm et al. 2010). To enhance sporulation, double-autoclaved pine needles were placed onto the surface of synthetic nutrient-poor agar medium (SNA; Nirenberg 1976). The cultures were incubated at 24 °C in the dark, microscopically examined after 2 wk and additionally inspected after 4 wk. Colony characters and pigment production were noted after 2 wk of growth on MEA, PDA and OA (Crous et al. 2009) incubated at 24 °C.
Species of Phomopsis were identified based on morphology of conidia formed in pycnidia (van Niekerk et al. 2005). The sporulation was enhanced by amending 2 % WA cultures with sterilised pine needles and incubating them at 25 °C under near UV light with a 12 h photoperiod. Isolates were examined weekly for formation of pycnidia and conidia.
Microscopic observations for all fungi were made from mycelium of colonies cultivated on the respective medium or by using slide culture technique, as explained by Arzanlou et al. (2007) when studying the genus Mycosphaerella. Photos were captured with a Nikon camera system (Digital Sight DXM 1200, Nikon Corp., Japan), with a Nikon SMZ1000 dissecting microscope (DM) or with a Nikon Eclipse 80i microscope using differential interference contrast illumination (DIC). Structures were mounted in lactic acid, and 30 measurements (1 000× magnification) were determined. The 5th and 95th percentiles were defined for all measurements with the extremes given in parentheses. Colony colours were determined using the colour charts of Rayner (1970). Cardinal growth temperatures were determined by incubating MEA plates in the dark at 6–40 °C with 3 °C intervals (Collophora) or 5–40 °C with 5 °C intervals (Phaeoacremonium), also including 37 °C. Radial growth was measured after 8 d at 25 °C (Phaeoacremonium) or 2 wk at 24 °C (Collophora). Isolates of novel species were deposited in the culture collection of the CBS-KNAW Fungal Biodiversity Centre (CBS), Utrecht, The Netherlands.
DNA isolation
Fungal mycelium and conidia from pure cultures grown on PDA for 2 wk at 25 °C in the dark were scraped and mechanically disrupted by grinding to a fine powder in liquid nitrogen with a mortar and pestle. Total genomic DNA was extracted with the E.Z.N.A. Plant Miniprep Kit (Omega Biotek, Norcross, GA) following the manufacturer’s instructions. DNA was visualised on 0.7 % agarose gels stained with ethidium bromide and the DNA aliquots were stored at −20 °C.
Molecular identification and phylogenetic analysis
Morphological identifications of Botryosphaeriaceae spp., diatrypaceous fungi and Phomopsis spp. were confirmed by sequence analysis of the internal transcribed spacer (ITS) nrDNA region using the primers ITS1 and ITS4 (White et al. 1990). Species in the Botryosphaeriaceae were also confirmed by analysis of partial β-tubulin gene (BT) sequences amplified using primers Bt2a and Bt2b (Glass & Donaldson 1995). For species of Phaeoacremonium, ± 600 bp of the 5′ end of the BT and ± 300 bp of the 5′ end of the actin (ACT) genes were amplified as described by Mostert et al. (2006) using primer sets T1 (O’Donnell & Cigelnik 1997) and Bt2b, and ACT-512F and ACT-783R (Carbone & Kohn 1999), respectively. For Collophora spp., the ITS region was amplified using the primer pairs ITS1-F (Gardes & Bruns 1993) and ITS4. Additionally, a 200-bp intron of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and a partial sequence of the translation elongation factor 1α (EF-1α) were amplified and sequenced using the primer pairs GDF1 and GDR1 (Guerber et al. 2003) and EF1-728F and EF1-986R (Carbone & Kohn 1999).
PCR products were purified with the High Pure PCR Product Purification Kit (Roche Diagnostics, Germany) and sequenced in both directions by Macrogen Inc., Sequencing Center (Seoul, South Korea). Sequences were edited using the Sequencher software v. 4.7 (Gene Codes Corporation, Ann Arbor, MI).
The Collophora sequences (ITS, GAPDH, EF-1α) were added to reference sequences (Damm et al. 2010) and the outgroup (Cadophora luteo-olivacea CBS 141.41, ITS: GU128588, GAPDH: JN808849, EF-1α: JN808856). The multi-locus alignment was manually adjusted using Sequence Alignment Editor v. 2.0a11 (Rambaut 2002). To determine whether the three sequence datasets were congruent and combinable, tree topologies of 70 % reciprocal Neighbour-Joining bootstrap with Maximum Likelihood distances (10 000 replicates) with substitution models determined separately for each partition using Modeltest v. 3.5 (Posada & Crandall 1998) were compared visually (Mason-Gamer & Kellogg 1996). The Phaeoacremonium sequences (BT and ACT) together with the reference sequences (Mostert et al. 2006, Damm et al. 2008b, Essakhi et al. 2008, Graham et al. 2009, Gramaje et al. 2009b) and the outgroup taxa, Pleurostomophora richardsiae (ACT: AY579271, BT: AY579334) and Wuestneia molokaiensis (ACT: AY579272, BT: AY579335) obtained from GenBank were aligned using MAFFT sequence alignment program v. 6 (Katoh & Toh 2010) followed by manual adjustments of the alignments in Sequence Alignment Editor v. 2.0a11. The BT and ACT alignments were concatenated. A partition homogeneity test was conducted in PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003). The congruence between the ACT and TUB datasets were tested at 1 000 replicates. Phylogenetic analyses of all aligned sequence data were performed with PAUP. Alignment gaps were treated as missing data and all characters were unordered and of equal weight. Any ties were broken randomly when encountered. All characters were unordered and of equal weight.
Maximum parsimony analysis was performed for the combined Phaeoacremonium dataset using the heuristic search option with 10 random simple taxon additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm with the option of saving no more than 10 trees with a score greater than or equal to 5 (Harrison & Langdale 2006). The maximum parsimony analysis for the combined Collophora dataset was performed using the heuristic search option with 100 random sequence additions and TBR without further restrictions for tree saving. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. The robustness of the trees obtained was evaluated by 1 000 bootstrap replications (Hillis & Bull 1993). Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were calculated.
Sequences derived in this study were lodged at GenBank, the alignments in TreeBASE (www.treebase.org/), and taxonomic novelties in MycoBank (www.MycoBank.org; Crous et al. 2004). GenBank accession numbers of the strains collected during this study are listed in Table 2. Additional GAPDH and EF-1α sequences were generated for strains CBS 141.41 (see above), CBS 120878 (JN808847, JN808854) and CBS 120873 (JN808848, JN808855).
RESULTS
Morphological identification and characterisation
Based on their appearance in culture, the isolates obtained in this study, could be assigned to five main fungal groups (Table 2). The first group was characterised by dark green or grey to dark grey fast-growing mycelium on PDA. Some isolates produced a yellow pigment after 3 days that diffuses into the agar. With age, most of these cultures developed single or grouped, black, globose fruiting bodies (pycnidia) on the surface of pine needles on WA releasing either pigmented or hyaline conidia. Based on descriptions of species of Botryosphaeriaceae (van Niekerk et al. 2004, Phillips 2006) and comparison with previously identified isolates from Spain, fungal cultures with pigmented conidia were assigned to two species: Diplodia seriata and Diplodia sp. Fungal cultures with hyaline conidia were assigned to three different species: Botryosphaeria dothidea, Neofusicoccum parvum and Neofusicoccum sp.
The second group of isolates was characterised by white to reddish cream, slow growing mycelium, turning red to blood colour with age. Isolates formed a red pigment that coloured the colony and surrounding medium. Conidiophores on hyphae were reduced to conidiogenous cells. Sporulation was abundant, with hyaline, 1-celled conidia aggregated in masses around the hyphae. All morphological characters observed were consistent with the description of Collophora spp. (Damm et al. 2010). However, the isolates could not be assigned to one of the known species.
The third fungal group was characterised by having white to white-cream cottony slow-growing mycelium on PDA lacking fruiting structures after an incubation time of 3 wk in the dark. With age, some cultures developed a grey pigment. After 3–4 wk under continuous fluorescent light, small black pycnidia were formed on the pine needles. Conidia developing in the fruiting bodies were filiform and mostly curved in shape, which corresponds to descriptions of species in the Diatrypaceae family (Glawe & Rogers 1984).
The fourth fungal group was characterised by pale to medium brown flat slow-growing cultures on MEA. Different types of phialides that were variable in size and shape were observed in the aerial mycelium, and either discrete or integrated in conidiophores. Sporulation was abundant and conidia hyaline and aseptate. All morphological characters corresponded to the genus Phaeoacremonium (Mostert et al. 2006).
The last fungal group was characterised by white, cottony, slow-growing, raised mycelium, with margins becoming pale brown with age. Dark brown, eustromatic pycnidia released a mucilaginous light-cream drop containing only one characteristic spore type, usually ovoid-ellipsoidal with one obtuse and one acute end. These morphological characteristics resembled those of Phomopsis species (van Niekerk et al. 2005).
Botryosphaeriaceae spp. were the most common fungi isolated from symptomatic almond wood from Mallorca Island, followed by Eutypa lata, Phaeoacremonium spp., Collophora hispanica and Phomopsis amygdali (Table 2). Of the Botryosphaeriaceae species isolated, N. parvum was the most abundant species (13 strains). In contrast, B. dothidea was the least abundant, with only one strain. Other species of Botryosphaeriaceae were also frequently isolated, and included D. olivarum (7 strains), N. australe (6 strains) and D. seriata (4 strains). Diatrypaceae, represented by E. lata, were also frequently isolated in this study (7 strains), while only one strain of Ps. amygdali was collected. While Pm. iranianum was infrequently isolated (one strain), the novel species Pm. amygdalinum and Co. hispanica were collected several times (4 strains each).
Species of Botryosphaeriaceae, Eutypa lata and Ps. amygdali isolates were mostly isolated from circular (Fig. 1c, g) or sectorial necrosis (Fig. 1f), and wood discoloration (Fig. 1e). Phaeoacremonium and Collophora spp. were isolated from black spots and dark brown to black streaking of the xylem tissue (Fig. 1d, h).
Molecular identification and phylogenetic analyses
To confirm the identification based on morphology, BLASTn searches in GenBank showed that ITS sequences of Botryosphaeriaceae isolates had 99–100 % identity with isolates of D. seriata CBS 121485 (GenBank EU650671), B. dothidea CBS 121484 (GenBank EU650670) and N. parvum CBS 110301 (GenBank AY259098). Diplodia sp. isolates showed 100 % identity with isolates previously described as D. olivarum (GenBank GQ923873, GQ923874; Lazzizera et al. 2008a), while Neofusicoccum sp. isolates showed 99 % identity with isolates previously identified as Neofusicoccum australe CBS 115185 (GenBank FJ150696) and CBS 119046 (GenBank DQ299244).
The ITS sequences of the second group of isolates were 96 % identical to those of Co. africana STE-U 6113 (GenBank GQ154570) and Co. capensis STE-U 6341 (GenBank GQ154574), while the EF sequences of these isolates were 91 % identical to that of Co. rubra STE-U 6198 (GenBank GQ154642). The ITS sequences of the Diatrypaceae isolates from this study had 99–100 % identity with isolates previously identified as Eutypa lata (GenBank AY462541, AY662394). Regarding Phaeoacremonium isolate Pir-1 from Mallorca Island, BT sequences had 99–100 % identity with Pm. iranianum isolates (GenBank EU128077, FJ872406). The ACT sequences of the remaining Phaeoacremonium isolates were 87 % identical to those of Pm. pallidum STE-U 6104 (GenBank EU128103) and Pm. viticola STE-U 6180 (GenBank EU128094), while the BT sequences were 81 % identical to those of Pm. theobromatis CBS 111586 (GenBank DQ173132) and Pm. viticola CBS 100947 (GenBank DQ173134). A BLASTn search showed that the ITS sequence of isolate Pam-1 had 100 % identity with isolates previously identified as Ps. amygdali (GenBank AF102996, AF102997). Sequences of three representative isolates of each species derived in this study were lodged in GenBank (Table 2).
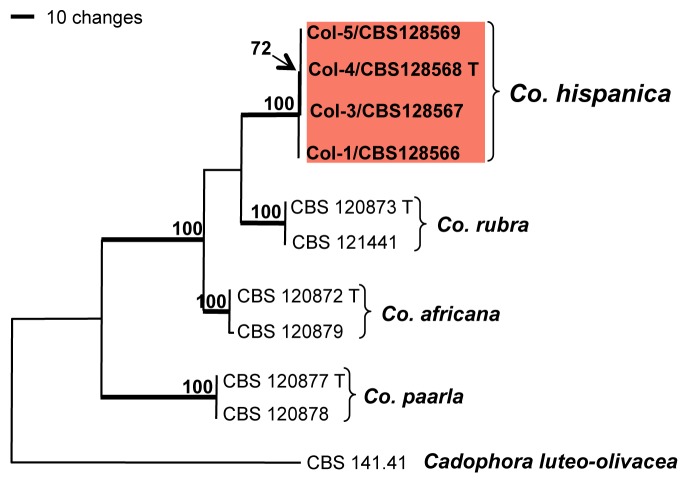
Phylogenetic analysis was performed only with genera of unknown species, Collophora and Phaeoacremonium (Fig. 2, 3). The ITS, GAPDH and EF-1α sequence datasets of the genus Collophora did not show any conflicts in tree topology for the 70 % reciprocal bootstrap trees, which allowed us to combine them. The combined sequence dataset consisted of 11 isolates including the outgroup and had 1 097 included characters, of which 133 characters were parsimony-informative, 130 parsimony-uninformative and 834 constant. After a heuristic search, two equally most parsimonious trees with identical topologies were retained (length = 318 steps, CI = 0.921, RI = 0.907, RC = 0.836) of which one is shown in Fig. 2. Isolates of Col-1, Col-3, Col-4 and Col-5 from almond trees on the island of Mallorca form a distinct clade (100 % bootstrap support) sister to Co. rubra. Sequences of the type strains of Co. africana (CBS 120872) and Co. capensis (CBS 120879) as well as Co. paarla (CBS 120877) and Co. pallida (CBS 120878) are each present in well-supported clades (100 % bootstrap support) with no or little variability.
Fig. 2.
One of two most parsimonious trees obtained from heuristic searches of ITS, GAPDH and EF-1α gene sequences of Collophora species. Bootstrap support (1 000 replicates) above 70 % are shown at the nodes. Cadophora luteo-olivacea CBS 141.41 was used as outgroup. Ex-type strains for each species are indicated with a ‘T’ after the strain number.
Fig. 3.
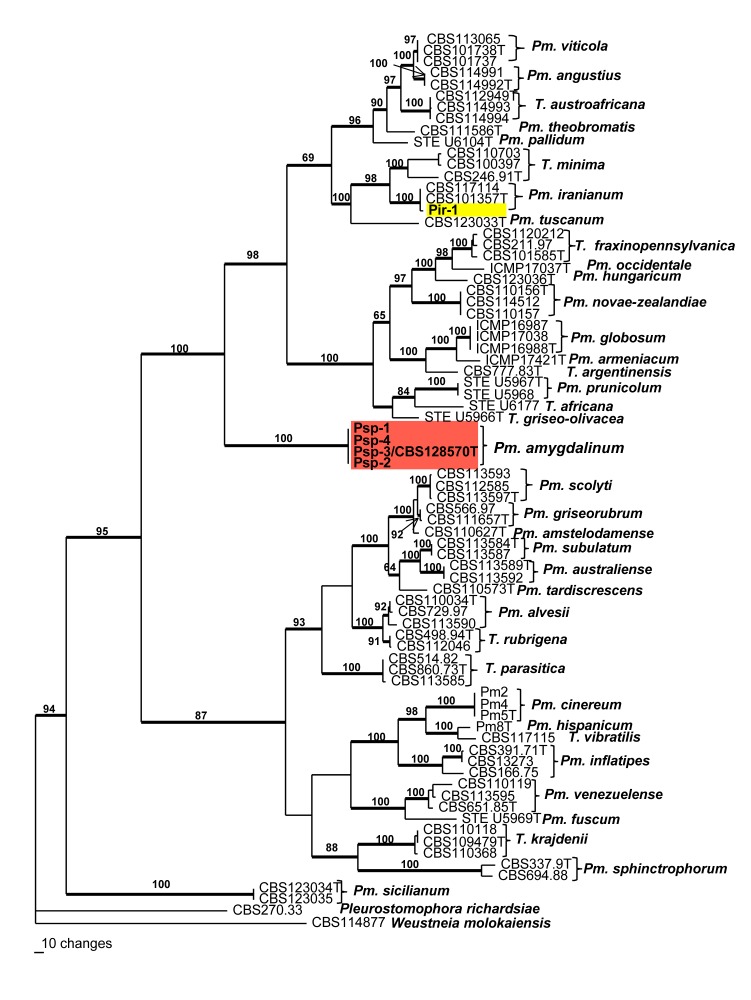
One of 90 most parsimonious trees obtained from heuristic searches of a combined alignment of the TB and ACT gene sequences. Bootstrap support (1 000 replicates) above 60 % are shown at the nodes. Pleurostomophora richardsiae and Wuestneia molokaiensis were used as outgroup. Ex-type strains for each species are indicated with a ‘T’ after the strain number. Thickened lines indicate branches present on strict consensus tree.
The partition homogeneity test of the BT and ACT alignments of Phaeoacremonium gave a P-value of 0.05 indicating that the datasets were congruent and could be combined. The combined sequence dataset consisted of 78 isolates including the outgroup and had 918 characters, of which 510 characters were parsimony-informative, 126 parsimony-uninformative and 282 constant. Ninety equally most parsimonious trees were retained (length = 2 690 steps, CI = 0.465, RI = 0.823, RC = 0.383). A tree that closely resembled the strict consensus tree was chosen and is presented in Fig. 3. The strain Pir-1, grouped inside the Pm. iranianum clade with 100 % bootstrap support. Four strains (Psp 1–4) grouped together in a monophyletic clade with 100 % bootstrap support, basal to the clades containing T. minima and Pm. novaezealandiae, with no other closely related species.
Taxonomy
Based on the DNA sequence analyses and morphological characters, two species, one species each of Collophora and Phaeoacremonium, proved distinct from all known species, and are newly described below.
Collophora hispanica D. Gramaje, J. Armengol & Damm, sp. nov. — MycoBank MB561926; Fig. 4
Fig. 4.
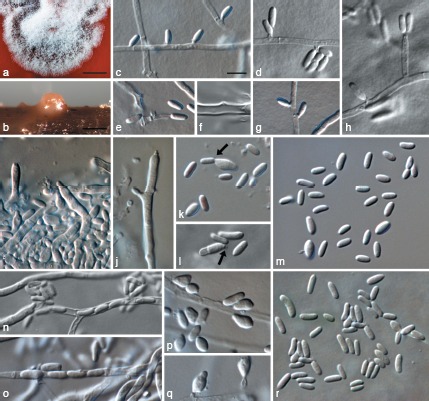
Collophora hispanica. a. Colony on MEA that is stained red by the pigment exuded by the fungus; b. conidioma on pine needle; c–h. conidiogenous cells and conidia on hyphal cells; i, j. conidiophores formed in conidiomata on pine needles; k, l. microcyclic conidiation (indicated by arrows) in conidia from conidiomata (k) and from hyphal cells (l); m. conidia formed in conidiomata after 4 wk; n, o. endoconidia; p, q. conidia formed on hyphal cells after 4 wk; r. conidia formed on hyphal cells after 2 wk. a, b: DIC, c–r: DM. — Scale bars: a = 1 mm; b = 100 μm; c = 5 μm; scale bar for c applies to c–r.
Etymology. Named after Spain where this fungus was first collected.
Vegetative hyphae hyaline, smooth-walled, septate, branched, 1–3.5 μm wide, lacking chlamydospores. Sporulation abundant, conidia formed on hyphal cells, occasionally in hyphae (endoconidia) and in conidiomata or due to microcyclic conidiation. Conidiophores on hyphae reduced to conidiogenous cells. Conidiogenous cells enteroblastic, hyaline, mainly intercalary, reduced to collarettes formed directly on hyphal cells or on short necks; necks cylindrical, 0.5–2 μm long, 0.5–1.5 μm wide; discrete phialides often observed, cylindrical to ampulliform, 4.5–8 × 1–2 μm; collarettes cylindrical to narrowly funnel shaped, very thin-walled, 0.5 μm long, opening 0.5 μm wide, inconspicuous, periclinal thickening sometimes visible. Conidia aggregated in masses around the hyphae, hyaline, 1-celled, cylindrical, sometimes obovate, often slightly bent, with both ends obtuse or with a papillate apex, smooth-walled, (2.5−)3.5–5(−6.5) × (1−)1.5(−2) μm, av. ± SD = 4.3 ± 0.7 × 1.5 ± 0.2 μm, L/W ratio = 2.8, after 4 wk many clavate, limoniform, subsphaerical or irregularly inflated conidia observed that are often > 2 μm wide. Microcyclic conidiation occurs occasionally, with conidia developing into mother cells, becoming > 7 μm long, 2–3 μm wide, and sometimes septate, with a short neck or with a mere opening with a minute collarette at one end. Endoconidia uniseriate within hyphae, hyaline, 1-celled, cylindrical, slightly bent, with both ends obtuse, same size as conidia formed on hyphal cells. Conidiomata occasionally formed on pine needles in 2–4 wk. Conidiophores hyaline to slightly reddish, smooth-walled, septate, branched. Conidiogenous cells enteroblastic, hyaline to slightly reddish, smooth-walled, cylindrical to ampulliform, conidiogenous loci formed terminal and intercalary, immediately below the septum (acropleurogenously), 3–6 × 1–3 μm, collarettes sometimes visible, ≤ 0.5 μm long, opening minute ≤ 0.5 μm wide, periclinal thickening not observed. Conidia hyaline or reddish, 1-celled, cylindrical, sometimes obovate, often slightly bent, with both ends obtuse or with a papillate apex, smooth-walled, (2.5−)3–5(−7) × 1–2(−3.5) μm, av. ± SD = 4.2 ± 1.0 × 1.7 ± 0.5 μm, L/W ratio = 2.5.
Culture characteristics — Colonies reaching a radius of 2–2.5 mm after 2 wk at 24 °C in the dark on MEA. Minimum temperature for growth < 6 °C, optimum 18 °C, maximum 30 °C. Colonies on PDA flat, moist, entire to undulate margin, coral, livid red to vinaceous with white to pale vinaceous margin, with little pale vinaceous to vinaceous aerial mycelium in the centre; reverse same colours, turning dark vinaceous with age, medium around culture vinaceous due to diffuse pigment; on OA flat to low convex, entire to slightly undulate margin, white to pale rosy vinaceous, surface covert almost entirely with white aerial mycelium, reverse salmon to flesh, white towards the margin; on MEA flat to low convex, undulate margin, dark vinaceous, surface covered partly or entirely with rosy to vinaceous aerial mycelium; reverse blood colour, medium around culture red due to diffusing pigment.
Specimens examined. Spain, Mallorca, Sant Llorenç del Cardassar, isolated from branches of Prunus dulcis trees, June 2010, J. Armengol, CBS H-20518 holotype, culture ex-type CBS 128568 = Col-4; Mallorca, Sant Llorenç del Cardassar, isolated from branches of Prunus dulcis trees, June 2010, J. Armengol, Col-1 herb, CBS H-20516, culture Col-1 = CBS 128566; Mallorca, Sant Llorenç del Cardassar, isolated from branches of Prunus dulcis trees, June 2010, J. Armengol, Col-3 herb, CBS H-20517, culture Col-3, CBS 128567; Mallorca, Sant Llorenç del Cardassar, isolated from branches of Prunus dulcis trees, June 2010, J. Armengol, Col-5 herb, CBS H-20519, culture Col-5, CBS 128569.
Notes — The phylogeny of the combined sequence dataset showed that Co. hispanica does not group with any of the known species.
Colonies of Co. hispanica resemble those of Co. africana and Co. rubra in forming red pigments that stain the colony and surrounding medium. Conidia formed in the mycelium are often slightly curved like those of Co. rubra. Unlike both of those species, discrete phialides are common and endoconidia are formed similar to Co. pallida and Co. paarla. Few conidiomata were formed on pine needles, but not on agar medium. The conidia in these conidiomata formed on conidiophores similar to those of other Collophora species such as Co. africana, however no wall structures were observed.
Collophora africana Damm & Crous, Persoonia 24: 65. 2010
= Collophora capensis Damm & Crous, Persoonia 24: 67. 2010.
Collophora paarla Damm & Crous, Persoonia 24: 67. 2010
= Collophora pallida Damm & Crous, Persoonia 24: 69. 2010.
Notes — Collophora africana and Co. capensis were regarded as distinct taxa based on differences in conidial morphology (on hyphae and in conidiomata) and differences in cardinal temperature requirements for growth. Furthermore, Co. pallida and Co. paarla were differentiated based on their conidial morphology, width of vegetative hyphae, and exudates formed in culture (Damm et al. 2010). However, the phylogeny of the multigene sequence dataset of Collophora spp. generated in this study (Fig. 2), only supports two species, namely Co. africana and Co. paarla, suggesting that the observed variation (Damm et al. 2010) was not informative at species level.
Phaeoacremonium amygdalinum D. Gramaje, J. Armengol & L. Mostert, sp. nov. — MycoBank MB561925; Fig. 5
Fig. 5.
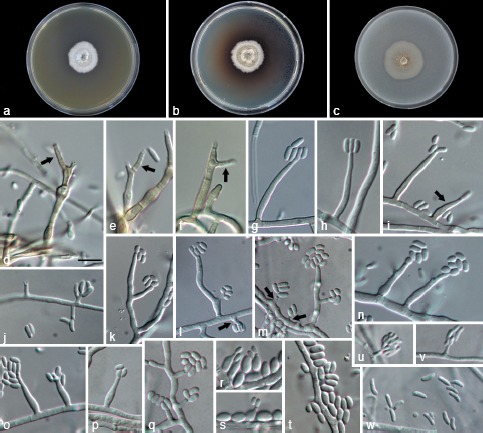
Phaeoacremonium amygdalinum. a–c. Sixteen-day-old colonies incubated at 25 °C on MEA (a), PDA (b) and OA (c); d–j. aerial structures on MEA; d–f. conidiophores with polyphialides (indicated by arrows); g, h. type III phialides; i. type III and type II phialides (indicated by arrow); j. type I phialides; k–t. aerial structures by using slide culture technique; k–m. branched conidiophores and type I phialides (indicated by arrows); n–p. type II phialides; q. type I phialides; r–s. Microcyclic conidiation; t. conidia; u–w. structures on the surface of and in MEA; u–v. adelophialides with conidia; w. conidia. — Scale bars: d = 10 μm; scale bar for d applies to d–w.
Etymology. Named after the host it was isolated from, almond (Prunus dulcis), which is in Greek amygdali (Αμυγδαλή).
Aerial structures in vitro on MEA: Mycelium consisting of branched, septate hyphae that occurs singly or in bundles of up to 10; hyphae tuberculate with warts up to 2 μm diam, verruculose to smooth, medium brown to pale brown and 2–3.5 μm wide. Conidiophores mostly short, usually unbranched, arising from aerial or submerged hyphae, erect to flexuous, up to 5-sepate, medium brown to pale brown, verruculose on the lower part, (12−)15.5–40(−55) (av. = 29) μm long and 1.5–3 (av. = 2.1) μm wide. Phialides terminal or lateral, often polyphialidic, smooth to verruculose, hyaline, collarettes, 1.5–2.5 μm long, 1–1.5 μm wide; type I phialides mostly cylindrical, occasionally widened at the base, (3−)3.5–7.5(−10) × 1–2 (av. = 6 × 1.5) μm; type II phialides most predominant, either subcylindrical or navicular, some elongate-ampulliform and attenuated at the base, (9−)10–16.5(−17) × 1.5–2.5 (av. = 13.5 × 2) μm; type III phialides cylindrical to subcylindrical, 17–27 × 1.5–2.5(−3) (av. = 21 × 2) μm. Conidia hyaline, oblong ellipsoidal or obovoid, (3−)4–5 × 1.5–3 (av. = 4.5 × 2) μm, L/W ratio = 2.1.
On surface or submerged in the agar — Phialides hyaline, mostly cylindrical, 4–12 × 1–2 (av. = 7.5 × 1.5) μm. Conidia hyaline, mostly allantoid, few reniform, (5−)5.5–7(−10) × 1–2 (av. = 6.5 × 1.5) μm, L/W ratio = 4.4.
Culture characteristics — Colonies reaching a radius of 9.5–10 mm after 8 d at 25 °C. Minimum temperature for growth 15 °C, optimum 25 °C, maximum 30–35 °C. Colonies on MEA flat, with entire margin; after 8 d and 16 d, white to brownish drab above, buff-yellow to yellowish olive in reverse; on PDA flat, felt-like with few woolly tufts near the centre, with entire margin; after 8 d and 16 d, white to olive-brown or olive-green above, pale brownish drab towards the edge to dark greyish brown in reverse; on OA flat, with entire margin; after 8 d and 16 d white to pale olive-grey above. Colonies producing pale brown pigment on PDA.
Specimens examined. Spain, Mallorca, Sant Llorenç del Cardassar, isolated from Prunus dulcis trees, June 2009, D. Gramaje, CBS H-20509 holotype, culture ex-type CBS 128570 = Psp-3; Mallorca, Sant Llorenç del Cardassar, isolated from Prunus dulcis trees, June 2009, D. Gramaje, Psp-1 herb, CBS H-20507, culture Psp-1; Mallorca, Sant Llorenç del Cardassar, isolated from Prunus dulcis trees, June 2009, D. Gramaje, Psp-2 herb, CBS H-20508, culture Psp-2; Mallorca, Sant Llorenç del Cardassar, isolated from Prunus dulcis trees, June 2009, D. Gramaje, Psp-4 herb, CBS H-20510, culture Psp-4.
Notes — The phylogeny of the combined sequence dataset showed that Pm. amygdalinum does not group with any of the known species. A distinguishing morphological feature is the frequent occurrence of polyphialides. Other Phaeoacremonium species that also form polyphialides include Pm. australiense, Pm. fuscum, Pm. krajdenii, Pm. occidentale, Pm. pallidum, Pm. prunicolum, Pm. scolyti, the anamorph of Togninia africana and the anamorph of T. griseo-olivacea. Phaeoacremonium amygdalinum can be distinguished from these species based on brown colonies, the production of pale brown pigment on PDA, and by the predominance of the type II phialides. This species often produced microcyclic conidia under slide culture conditions.
DISCUSSION
This study shows the high incidence and diversity of fungal trunk pathogens associated with wood decay symptoms on almond trees on the island of Mallorca. These include species of Botryosphaeriaceae, Eutypa lata, Phaeoacremonium iranianum, Phomopsis amygdali and the new species described here, namely Collophora hispanica and Phaeoacremonium amygdalinum. These species could be distinguished based on DNA sequence data and unique morphological characters.
Several species of Botryosphaeriaceae were isolated from wedge-shaped wood necroses on almond trees. The majority of Botryosphaeriaceae isolates belonged to Neofusicoccum australe, N. parvum and Diplodia olivarum, while Botryosphaeria dothidea and D. seriata were only occasionally isolated.
Botryosphaeria dothidea causes canker diseases in a broad range of woody plants, including several Prunus spp. (English et al. 1966, Sutton 1980). A canker of trunk and scaffold branches of almond trees was reported by English et al. (1966). This disease, sometimes called ‘band canker’, was first noted in 1959 occurring in several counties in California. However, some of these reports need to be viewed with care since many species have been relegated incorrectly to the name B. dothidea. For instance, N. ribis (as B. ribis) was previously regarded as a synonym of B. dothidea (von Arx & Müller 1954). Neofusicoccum parvum (as B. parva) was often not distinguished from N. ribis and consequently treated as B. dothidea (Slippers et al. 2004). In Spain, B. dothidea, together with D. seriata and N. parvum, is considered as the most common species associated with grapevine (Vitis vinifera) decline syndrome (Armengol et al. 2001, Aroca et al. 2006). Additionally, this species has recently been isolated from olive fruits in southern Spain showing symptoms of dalmatian disease (Moral et al. 2010). Botryosphaeria dothidea was not found during surveys on stone and pome fruit trees in South Africa (Damm et al. 2007a, Slippers et al. 2007), however it was confirmed to be associated with band canker of almond trees in California (Inderbitzin et al. 2010). This study also represents the first record of D. olivarum on almond. This species was recently associated with diseased olive drupes (Lazzizera et al. 2008a) and carob trees (Ceratonia siliqua) (Granata et al. 2011) in Italy.
The low incidence of D. seriata agrees with the results of Inderbitzin et al. (2010) from almond and peach trees in California. In contrast, this species was the most frequently isolated Botryosphaeriaceae species (43 of 67 isolates) on apricot, nectarine, peach and Japanese plum in South Africa (Damm et al. 2007a). It was also the dominant species in a study on stone and pome fruit trees in South Africa by Slippers et al. (2007), which represented over 90 % of the isolates collected over a 5-year period and in a recent study on pome fruit trees in South Africa by Cloete et al. (2011). Diplodia seriata is known to occur on a wide range of hosts (Punithalingam & Waller 1973) and to cause severe diseases in some host plants, such as apple (Malus domestica) or peach (Britton & Hendrix 1982). Farr et al. (2008) listed 264 hosts under its former name of Botryosphaeria obtusa. In Spain, D. seriata also occurs on olive drupes (Moral et al. 2007).
Neofusicoccum australe was frequently isolated from almond trees studied here. This species was reported from almond and plum in the Western Cape, South Africa, by Slippers et al. (2007) who considered it to be infrequent and of minimal importance on stone fruits. However, Damm et al. (2007a) found N. australe commonly on three Prunus species (peach, Japanese plum and apricot) and in different locations in this region of South Africa. This fungus is the dominant Botryosphaeriaceae species infecting native Eucalyptus species in Western Australia (Burgess et al. 2005). Neofusicoccum australe was recently reported from Eucalyptus and pistachio (Pistacia vera) trees in Spain (Armengol et al. 2008), from olives in Italy (Lazzizera et al. 2008b), from avocado (Persea americana) in California (McDonald et al. 2009) and from grapevines in Australia (Taylor et al. 2005), New Zealand (Amponsah et al. 2009), South Africa (van Niekerk et al. 2004) and Spain (Aroca et al. 2010).
Neofusicoccum parvum was the most frequently isolated Botryosphaeriaceae species in this study. Recently, this fungus was reported affecting almond trees in California (Inderbitzin et al. 2010). Neofusicoccum parvum is a common pathogen of pome and stone fruit trees world-wide (Slippers et al. 2007), and judging from the frequency of isolation, it seems to be one of the most common causes of wood decay of almond trees in Spain. Therefore, it should also be taken into account with the development of disease control measures. In Spain, N. parvum has also been isolated from English walnut (Juglans regia) and Japanese plum trees (Moral et al. 2010).
Several strains of Eutypa lata were isolated during this study. This fungus is a major pathogen of cultivated crops such as apricot and grapevine and has been found all over the world (Carter 1957). In almond, the occurrence of a perithecial stroma on the dead stump of a tree was first reported by Carter (1960) in Australia. This species has also been recorded from necrotic vascular tissue associated with cankers in almond trees in Greece (Carter 1982, Rumbos 1985).
Phaeoacremonium iranianum has previously been reported affecting kiwifruit in Italy (Mostert et al. 2006) and grapevines in several countries, such as Italy (Essakhi et al. 2008), Iran (Mostert et al. 2006) and Spain (Gramaje et al. 2009a). This is the first report of Pm. iranianum on almond. This species was recently isolated from necrotic wood of apricot in South Africa (Damm et al. 2008b). However, the impact of Pm. iranianum on dieback disease on almond trees in Spain is uncertain, since only one isolate was obtained. In contrast, the new species, Pm. amygdalinum was found to be more frequently associated with wood decay symptoms of almond trees during this study.
Phomopsis amygdali was isolated from affected shoots of almond trees. This species was recently reported affecting almond branches in Portugal (Diogo et al. 2010) and Tunisia (Rhouma et al. 2008). However, Ps. amygdali is not restricted to Prunus spp. but has also been isolated from grapevines in South Africa (Mostert et al. 2001).
The fungi reported in this study were isolated from necrotic wood tissue of almond trees on the island of Mallorca. We did not determine their pathogenicity, but, most of these fungi had previously been reported to be pathogenic or potentially pathogenic to Prunus spp., such as B. dothidea (English et al. 1966, Inderbitzin et al. 2010), D. seriata (Britton & Hendrix 1982, Britton et al. 1990, Damm et al. 2007a, Inderbitzin et al. 2010), N. australe (Damm et al. 2007a), N. parvum (Inderbitzin et al. 2010), Eutypa lata (Carter & Moller 1971, English & Davis 1978, Carter 1982, Rumbos 1985), Pm. iranianum (Damm et al. 2008b) and Ps. amygdali (Diogo et al. 2010).
The results of the isolations made during this study show the complex situation generated by many fungal trunk pathogens on almond trees on the island of Mallorca, which is in agreement with previous reports on almond and several Prunus spp. in other regions in the world (Damm et al. 2007a, 2008b, 2009, 2010, Slippers et al. 2007, Inderbitzin et al. 2010).
In Spain, as well as in other countries, commercial Prunus orchards are often planted adjacent to vineyards. Most of the species isolated from almond trees in this study are known grapevine pathogens in different regions of the world. For example, species of Botryosphaeriaceae are important pathogens of grapevine, causing cankers and other dieback symptoms in all major viticulture regions worldwide (van Niekerk et al. 2004). Eutypa lata is the causal agent of eutypa dieback, an important perennial canker disease that occurs in most countries where grapevine is cultivated (Munkvold et al. 1994). Species of Phaeoacremonium have been associated with very destructive grapevine decline diseases such as Petri disease and esca (Mostert et al. 2006). Phomopsis cane and leaf spot is an important disease of grapevines, causing serious losses to the wine industry (Mostert et al. 2001). Therefore, these fungal species could have spread from grapevine plants to almond trees. Conversely, almond orchards should be considered as potential sources of viable inoculum for trunk disease pathogens from which grapevines could be infected and almond trees could serve as an additional mode of pathogen survival in the absence of grapevine plants. Further studies on the adjacent stands to the ones sampled here could provide new insights on the epidemiology of fungal trunk pathogens.
Disease management practices employed on farms where vine-yards are planted in close proximity to Prunus orchards are therefore crucial for disease prevention. Since these trunk pathogens mainly infect fresh wounds such as pruning wounds via air- and waterborne inoculum (Trese et al. 1980, Hewitt & Pearson 1988, Pscheidt & Pearson 1989, Larignon & Dubos 2000), having a low inoculum pressure would be a logical condition for preventing infections. Methods by which the infection of wounds could be prevented or at least reduced include the removal of dead wood from vineyard or Prunus orchard floors, in order to minimise possible infection sources, avoiding pruning immediately following rainfall and applying fungicides or biological control agents to wound surfaces immediately after pruning.
Acknowledgments
We thank ‘Conselleria d’Agricultura, Mediambient i Territori’ of the Balearic Islands for funding this research. We appreciate the helpful assistance in field works from technicians and land owners: Tomeu Melis, Toni Nicolau, Beatriz Blanquer and Alejandro Aristondo. The authors also wish to thank the staff of ‘Conselleria d’Agricultura, Mediambient i Territori’: Alicia Nieto, Xavier Nadal and Toni Martorell. We would like to thank Walter Gams for kindly assisting with the nomenclature for the new species.
REFERENCES
- Agarwal DK, Chowdhry PN, Sarbhoy AK. 1992. Contributions to our knowledge of Aplosporella. Mycotaxon 45: 389–403 [Google Scholar]
- Amponsah NT, Jones EE, Ridgway HJ, Jaspers MV. 2009. First report of Neofusicoccum australe (Botryosphaeria australis), a cause of grapevine dieback in New Zealand. Australasian Plant Disease Notes 4: 6–8 [Google Scholar]
- Armengol J, Gramaje D, Pérez-Sierra A, Landeras E, Alzugaray R, Martos S, Luque J. 2008. First report of canker disease caused by Neofusicoccum australe on eucalyptus and pistachio in Spain. Plant Disease 92: 980. [DOI] [PubMed] [Google Scholar]
- Armengol J, Vicent A, Torn L, García-Figueres F, García-Jiménez J. 2001. Fungi associated with esca and grapevine declines in Spain: a three-year survey. Phytopathologia Mediterranea 40: S325–S329 [Google Scholar]
- Aroca A, Garcia-Figueres F, Bracamonte L, Luque J, Raposo R. 2006. A survey of trunk disease pathogens within rootstocks of grapevines in Spain. European Journal of Plant Pathology 115: 195–202 [Google Scholar]
- Aroca A, Gramaje D, Armengol J, García-Jiménez J, Raposo R. 2010. Evaluation of grapevine nursery process as a source of Phaeoacremonium spp. and Phaeomoniella chlamydospora and occurrence of trunk disease pathogens in rootstock mother vines in Spain. European Journal of Plant Pathology 126: 165–174 [Google Scholar]
- Arx JA von, Müller E. 1954. Die Gattungen der amerosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz 11: 1–434 [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton KO, Hendrix FF. 1982. Three species of Botryosphaeria cause peach tree gummosis in Georgia. Plant Disease 66: 1120–1121 [Google Scholar]
- Britton KO, Hendrix FF, Pusey PL, Okie WR, Reilly CC, Daniell JW. 1990. Evaluating the reaction of peach cultivars to infection by three Botryosphaeria species. HortScience 25: 468–470 [Google Scholar]
- Burgess TI, Barber PA, Hardy GEStJ. 2005. Botryosphaeria spp. associated with eucalypts in Western Australia, including the description of Fusicoccum macroclavatum sp. nov. Australasian Plant Pathology 34: 557–567 [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556 [Google Scholar]
- Carter MV. 1957. Eutypa armeniacae Hansf. & Carter, sp. nov., an airborne vascular pathogen of Prunus armeniaca L. in southern Australia. Australian Journal of Botany 5: 21–35 [Google Scholar]
- Carter MV. 1960. Further studies on Eutypa armeniacae Hansf. & Carter. Australian Journal of Agricultural Research 11: 498–504 [Google Scholar]
- Carter MV. 1982. Additional hosts of Eutypa armeniacae in Australia. Australasian Plant Pathology 11: 46–48 [Google Scholar]
- Carter MV, Moller MJ. 1971. The quantity of inoculum required to infect apricot and other Prunus species with Eutypa armeniacae. Australian Journal of Experimental Agriculture 11: 684–686 [Google Scholar]
- Cloete M, Fourie PH, Damm U, Crous PW, Mostert L. 2011. Fungi associated with die-back symptoms of apple and pear trees with a special reference to grapevine trunk disease pathogens. Phytopathologia Mediterranea 50 (supplement): 176–190 [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, Samson RA. (eds). 2009. Fungal Biodiversity. CBS Laboratory Manual Series 1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2007a. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia 99: 664–680 [DOI] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2008a. A bitunicate ascus mechanism in the Calosphaeriaceae, with novel species of Jattaea and Calosphaeria on Prunus wood. Persoonia 20: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2009. Stone fruit trees – an inoculum source of grapevine trunk disease pathogens? Phytopathologia Mediterranea 48: 175–176 [Google Scholar]
- Damm U, Fourie PH, Crous PW. 2007b. Aplosporella prunicola, a novel species of anamorphic Botryosphaeriaceae. Fungal Diversity 27: 35–43 [Google Scholar]
- Damm U, Fourie PH, Crous PW. 2010. Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 24: 60–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Mostert L, Crous PW, Fourie PH. 2008b. Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 20: 87–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Verkley GJM, Crous PW, Fourie PH, Haegi A, Riccioni L. 2008c. Paraconiothyrium species on stone fruit trees and other woody hosts, including P. variabile and P. africanum spp. nov. Persoonia 20: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra OD, Sinclair JB. 1995. Basic plant pathology methods. 2nd edn CRC Press, Boca Raton, FL, USA [Google Scholar]
- Diogo ELF, Santos JM, Phillips AJL. 2010. Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115 [Google Scholar]
- Ellis MB, Ellis JP. 1997. Microfungi on land plants: An identification handbook. The Richmond Publishing Co., Ltd [Google Scholar]
- English H, Davis JR. 1965. Apricot dieback fungus found on western chokecherry. Plant Disease Reports 49: 178 [Google Scholar]
- English H, Davis JR. 1978. Eutypa armeniacae in apricot: pathogenesis and induction of xylem soft rot. Hilgardia 46: 193–204 [Google Scholar]
- English H, Davis JR, deVay JE. 1966. Dothiorella canker, a new disease of almond trees in California. Phytopathology 56: 146 [Google Scholar]
- Essakhi S, Mugnai L, Crous PW, Groenewald JZ, Surico G. 2008. Molecular and phenotypic characterization of novel Phaeoacremonium species associated with Petri disease and esca of grapevine. Persoonia 21: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2010. Food and Agriculture Organization of the United Nations, Statistical Databases. Available at http://www.fao.org, accessed on 25 September 2010. [Google Scholar]
- Farr DF, Bills GF, Chamuris GP, Rossman AY. 1989. Fungi on plants and plant products in the United States. St Paul, American Phytopathological Society [Google Scholar]
- Farr DF, Rossman AY, Palm ME, McCray EB. 2008. Fungal databases, systematic botany & mycology laboratory, ARS, USDA. Retrieved January 17, 2008, from http://nt.ars-grin.gov/fungaldatabases/ [Google Scholar]
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous infection due to Phaeoacremonium spp. Journal of Clinical Microbiology 41: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe DA, Rogers JD. 1984. Diatrypaceae in the Pacific Northwest. Mycotaxon 20: 401–460 [Google Scholar]
- Graham AB, Johnston PR, Weir BS. 2009. Three new Phaeoacremonium species on grapevines in New Zealand. Australasian Plant Pathology 38: 505–513 [Google Scholar]
- Gramaje D, Armengol J, Colino M, Santiago R, Moralejo E, Olmo D, Luque J, Mostert L. 2009a. First report of Phaeoacremonium inflatipes, P. iranianum and P. sicilianum causing Petri disease of grapevine in Spain. Plant Disease 93: 964. [DOI] [PubMed] [Google Scholar]
- Gramaje D, Armengol J, Mohammadi H, Banihashemi Z, Mostert L. 2009b. Novel Phaeoacremonium species associated with Petri disease and esca of grapevines in Iran and Spain. Mycologia 101: 920–929 [DOI] [PubMed] [Google Scholar]
- Granata G, Faedda R, Sidoti A. 2011. First report of canker disease caused by Diplodia olivarum on carob tree in Italy. Plant Disease 95: 776. [DOI] [PubMed] [Google Scholar]
- Grove WB. 1935. British stem- and leaf-fungi: coelomycetes vol. 1. Sphaeropsidales with hyaline conidia. Cambridge University Press (CUP) [Google Scholar]
- Guerber JC, Liu B, Correll JC, Johnston PR. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872–895 [PubMed] [Google Scholar]
- Gure A, Slippers B, Stenlid A. 2005. Seed-borne Botryosphaeria spp. from native Prunus and Podocarpus trees in Ethiopia, with a description of the anamorph Diplodia rosulata sp. nov. Mycological Research 109: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Harrison JC, Langdale JA. 2006. A step by step guide to phylogeny reconstruction. Plant Journal 45: 561–572 [DOI] [PubMed] [Google Scholar]
- Hausner G, Eyjólfsdóttir GG, Reid J, Klassen GR. 1992. Two additional species of the genus Togninia. Canadian Journal of Botany 70: 724–734 [Google Scholar]
- Hawksworth DL, Gibson IAS, Gams W. 1976. Phialophora parasitica associated with disease conditions in various trees. Transactions of the British Mycological Society 66: 427–431 [Google Scholar]
- Hewitt WB, Pearson RC. 1988. Diplodia cane dieback and bunch rot. In: Pearson RC, Goheen AC. (eds), Compendium of grape diseases: 25–26. APS Press, MN, USA [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192 [Google Scholar]
- Inderbitzin P, Bostock RM, Trouillas FP, Michailides TJ. 2010. A six locus phylogeny reveals high species diversity in Botryosphaeriaceae from California almond. Mycologia 102: 1350–1368 [DOI] [PubMed] [Google Scholar]
- INE. 2011. Instituto Nacional de Estadística (National Statistics Institute). Available at http://www.ine.es, accessed on 4 October 2011. [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larignon P, Dubos B. 2000. Preliminary studies on the biology of Phaeoacremonium. Phytopathologia Mediterranea 39: 184–189 [Google Scholar]
- Laundon GF. 1973. Botryosphaeria obtusa, B. stevensii and Otthia spireae in New Zealand. Transactions of the British Mycological Society 61: 369–374 [Google Scholar]
- Lazzizera C, Frisullo S, Alves A, Lopes J, Phillips AJL. 2008a. Phylogeny and morphology of Diplodia species on olives in southern Italy and description of Diplodia olivarum sp. nov. Fungal Diversity 31: 63–71 [Google Scholar]
- Lazzizera C, Frisullo S, Alves A, Phillips AJL. 2008b. Morphology, phylogeny and pathogenicity of Botryosphaeria and Neofusicoccum species associated with drup rot in olives in southern Italy. Plant Pathology 57: 948–956 [Google Scholar]
- Mason-Gamer R, Kellogg E. 1996. Testing for phylogenetic conflict among datasets in the tribe Triticeae (Graminae). Systematic Biology 45: 524–545 [Google Scholar]
- McAlpine D. 1902. Fungus diseases of stone-fruit trees in Australia and their treatment. Agriculture Department of Victoria, Melbourne, Australia [Google Scholar]
- McDonald V, Lynch S, Eskalen A. 2009. First report of Neofusicoccum australe, N. luteum, and N. parvum associated with avocado branch canker in California. Plant Disease 93: 967. [DOI] [PubMed] [Google Scholar]
- Moral J, Luque F, Trapero A. 2007. First report of Diplodia seriata, the anamorph of “Botryosphaeria” obtusa, causing fruit rot of olive in Spain. Plant Disease 92: 311. [DOI] [PubMed] [Google Scholar]
- Moral J, Muñoz-Díez C, González N, Trapero A, Michailides TJ. 2010. Characterization and pathogenicity of Botryosphaeriaceae species collected from olives and other hosts in Spain and California. Phytopathology 100: 1340–1351 [DOI] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Groenewald JZ, Gams W, Summerbell RC. 2003. Togninia (Calosphaeriales) is confirmed as teleomorph of Phaeoacremonium by means of morphology, sexual compatibility, and DNA phylogeny. Mycologia 95: 646–659 [DOI] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang JC, Phillips AJL. 2001. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167 [Google Scholar]
- Mostert L, Groenewald JZ, Summerbell RC, Gams W, Crous PW. 2006. Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Studies in Mycology 54: 1–115 [Google Scholar]
- Munkvold GP, Duthie JA, Marois JJ. 1994. Reductions in yield and vegetative growth of grapevines due to Eutypa dieback. Phytopathology 84: 186–192 [Google Scholar]
- Munkvold GP, Marois JJ. 1994. Eutypa dieback of sweet cherry and occurrence of Eutypa lata perithecia in the central valley of California. Plant Disease 78: 200–207 [Google Scholar]
- Niekerk JM van, Crous PW, Groenewald JZ, Fourie PH, Halleen F. 2004. DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia 96: 781–798 [PubMed] [Google Scholar]
- Niekerk JM van, Groenewald JZ, Farr DF, Fourie PH, Halleen F, Crous PW. 2005. Reassessment of Phomopsis species on grapevine. Australasian Plant Pathology 34: 27–39 [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117 [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116 [DOI] [PubMed] [Google Scholar]
- Phillips AJL. 2006. The Botryosphaeria site. http://www.crem.fct.unl.pt/ botryosphaeria_site [Google Scholar]
- Popushoi IS. 1971. Microflora plodovykh derevyaev SSSR (Mycoflora of fruit trees in the USSR). Moscow. [Google Scholar]
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Pscheidt JW, Pearson RC. 1989. Effect of grapevine training systems and pruning practices on occurrence of Phomopsis cane and leaf spot. Plant Disease 73: 825–828 [Google Scholar]
- Punithalingam E, Waller JM. 1973. Botryosphaeria obtusa. CMI Descriptions of Pathogenic Fungi and Bacteria, No. 394. Commonwealth Mycological Institute, Kew, UK [Google Scholar]
- Pusey PL, Kitajima H, Wu Y. 1995. Fungal gummosis. In: Ogawa JM, Zehr EI, Bird GW, Ritchie DF, Uriu K, Uyemoto JK. (eds), Compendium of stone fruit diseases: 98. St Paul, APS Press [Google Scholar]
- Rambaut A. 2002. Sequence Alignment Editor. Version 2.0. University of Oxford, Oxford, UK [Google Scholar]
- Rayner RW. 1970. A mycological color chart. Commonwealth Mycological Institute and British Mycological Society, Kew, Surrey, United Kingdom [Google Scholar]
- Rhouma A, Triki MA, Ouerteni K, Mezghanni M. 2008. Chemical and biological control of Phomopsis amygdali the causal agent of constriction canker of almond in Tunisia. Tunisian Journal of Plant Protection 3: 69–77 [Google Scholar]
- Rumbos IC. 1985. Further pathogenicity studies of Eutypa lata (= E. armeniacae) on almond. Options Mediterraneennes 1: 79–89 [Google Scholar]
- Rumbos IC. 1986. Phialophora parasitica, causal agent of cherry dieback. Journal of Phytopathology 117: 283–287 [Google Scholar]
- Slippers B, Crous PW, Denman S, Coutinho TA, Wingfield BD, Wingfield MJ. 2004. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96: 83–101 [PubMed] [Google Scholar]
- Slippers B, Smit WA, Crous PW, Coutinho TA, Wingfield BD, Wingfield MJ. 2007. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit trees in South African and other regions of the world. Plant Pathology 56: 128–139 [Google Scholar]
- Smit W, Viljoen C, Wingfield B, Wingfield M, Calitz F. 1996. A new canker disease of apple, pear, and plum rootstocks caused by Diaporthe ambigua in South Africa. Plant Disease 80: 1331–1335 [Google Scholar]
- Sutton BC. 1980. The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, MA, USA [Google Scholar]
- Taylor A, Hardy GEStJ, Wood P, Burgess T. 2005. Identification and pathogenicity of Botryosphaeria species associated with grapevine decline in Western Australia. Australasian Plant Pathology 34: 187–195 [Google Scholar]
- Trese AT, Burton CL, Ramsdell DC. 1980. Eutypa armeniacae in Michigan vineyards: Ascospore production and survival, host infection, and fungal growth at low temperatures. Phytopathology 70: 788–793 [Google Scholar]
- Trouillas FP, Úrbez-Torres JR, Gubler WD. 2010. Diversity of diatrypaceous fungi associated with grapevine canker disease in California. Mycologia 102: 319–336 [DOI] [PubMed] [Google Scholar]
- Tuset JJ, Portilla MT. 1989. Taxonomic status of Fusicoccum amygdali and Phomopsis amygdalina. Canadian Journal of Botany 67: 1275–1280 [Google Scholar]
- Uecker FA. 1988. A world list of Phomopsis names with notes on nomenclature, morphology, and biology. Mycologia Memoir No. 13. Cramer, Berlin [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungi ribosomal RNA genes for phylogenetics. In: PCR protocols. A guide to methods and applications: 315–322. Academic Press, San Diego, CA [Google Scholar]
- Wollenweber HW. 1941. Diplodia sarmentorum Fries und ihre Verbreitung. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene, Abteilung 2, 103: 347–357 [Google Scholar]
- Wollenweber HW, Hochapfel H. 1941. Beiträge zur Kenntnis parasitärer und saprophytischer Pilze 1. Diplodia und ihre Beziehung zur Fruchtfäule. Zeitschrift für Parasitenkunde 12: 167–250 [Google Scholar]