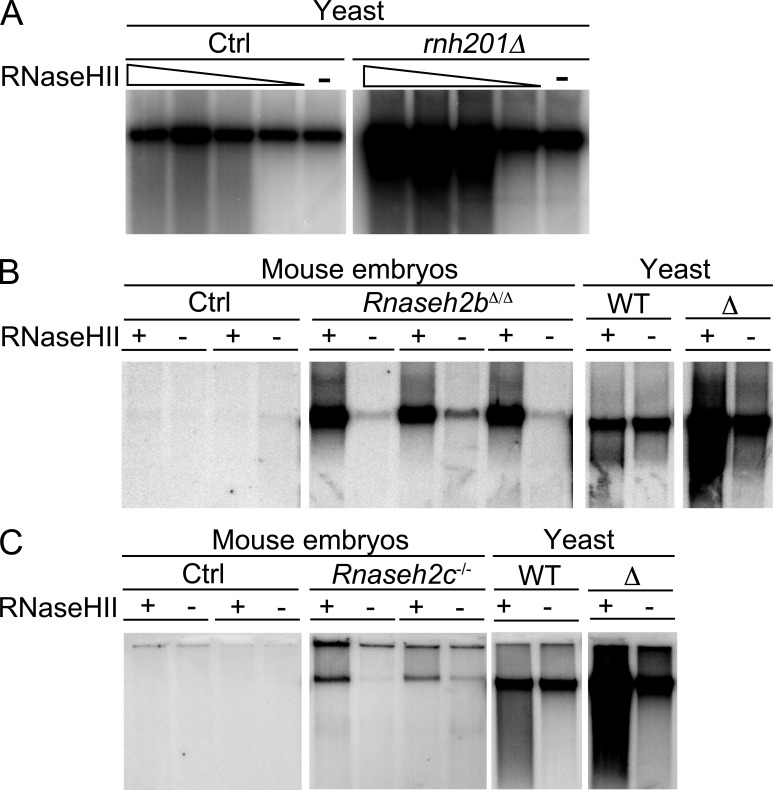
Figure 5.
Increased numbers of ribonucleotides in genomic DNA of RNase H2–deficient embryos. Large chromosomal fragments obtained by standard DNA purification were nicked by bacterial RNase HII at ribonucleotide positions, followed by DNA polymerase I–dependent nick translation in the presence of 32P-dCTP. Labeled DNA was run on a 1% agarose gel (focusing the large chromosomal fragments [several hundred kilobases] in a distinct band) and visualized by autoradiography. Control samples were not digested with RNase HII to detect background single strand breaks. (A) DNA from RNase H2–deficient (rnh201Δ) or control yeast (Ctrl) was digested with increasing amounts (1–10 mU/µl) of RNase HII or subjected to nick translation without RNase HII pretreatment. (B) 200 ng DNA from WT (Ctrl) and Rnaseh2bΔ/Δ embryos (obtained by crossing Rnaseh2bFLOX mice with PGK-Cre mice) was digested with 10 mU/µl RNase HII before nick translation. Negative control samples from each embryo were not pretreated with RNase HII. DNA from RNase H2–proficient (WT) and –deficient (Δ) yeast served as controls. (C) Experiment as in B performed with DNA from Rnaseh2c−/− and WT control embryos. All panels in A, B, and C originate from the same exposure of one single gel each.