Figure 3.
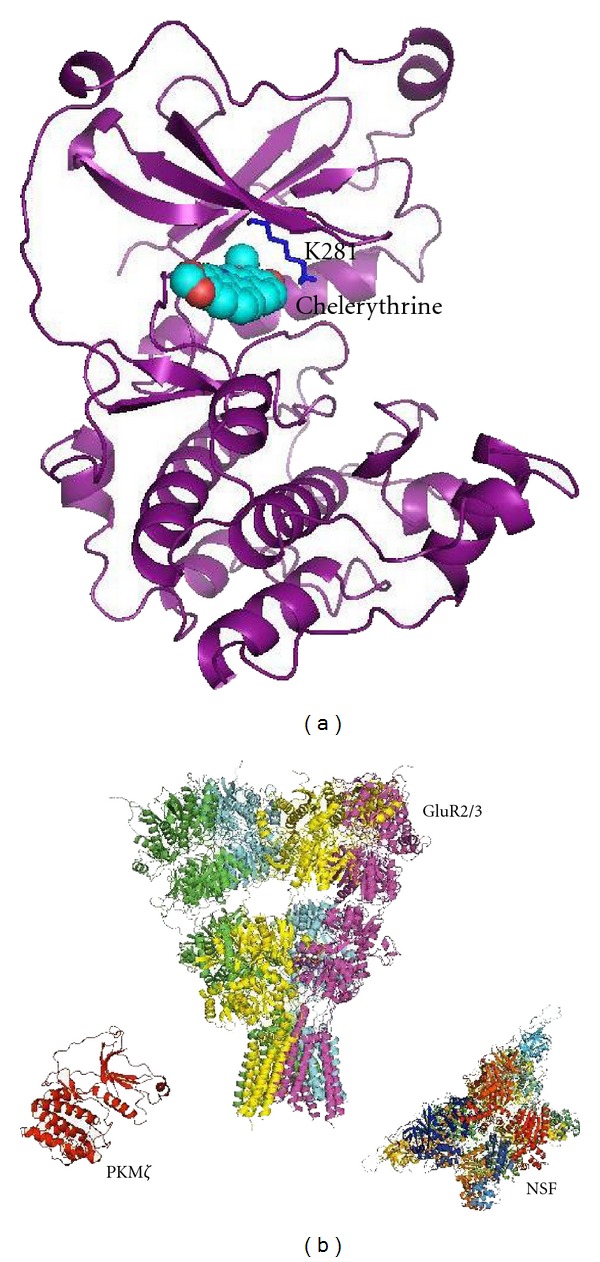
3D models of proteins involved in the long-term memory maintenance: (a) human PKMζ homology model based on PKC iota [1ZRZ.pdb]. The chelerythrine (sphere-filled model with the oxygen atoms shown as red spheres and carbon atoms as cyan) binding site was identified within the ATP-binding pocket of the PKMζ (ribbons). Docking was performed using AutoDock software [117] and protein was visualized using PyMol. The inhibitor binding site is located in the vicinity of the Lys281 residue. K281W mutation renders PKMζ inactive and causes permanent erasure of the long-term memory [118]. (b) The N-ethylmaleimide-sensitive factor (NSF) consists out of two hexameric Walker-type D1 and D2 domains (N-terminal NSF domain not shown). Its ATPase-catalytic activity is needed to release GluR2 subunit from endosomes and GluR2/3 assembly at the postsynaptic membrane. Crystallographic structure for the last 68 amino acids of the GluR2 C-terminal, including the NSF and AP2 binding sites, is not available.
