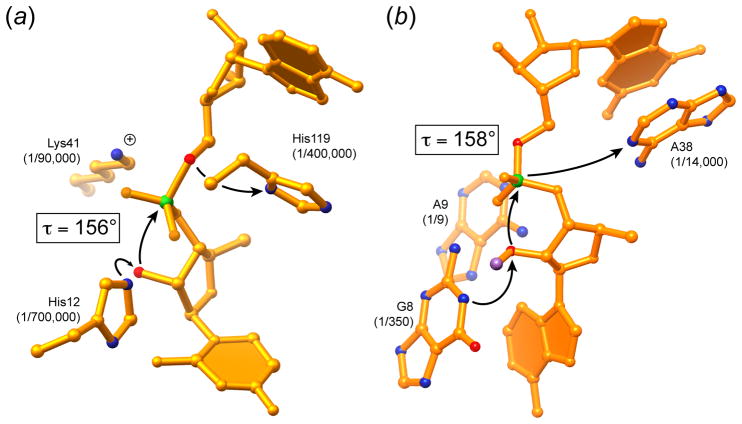
Figure 5.
Comparison of the active sites and mechanisms of RNase A and the hairpin ribozyme. (a) Active site of RNase A bound to the dinucleotide mimic inhibitor UpcA (Richards et al., 1971). In this compound, the atom corresponding to the 5′-oxo leaving group has been replaced with a methylene group. The degree of impairment of the enzyme when the catalytic histidines are replaced with alanines, or the catalytic lysine with cysteine (Raines, 1998) is indicated. τ is the angle formed between the nucleophile, electrophile and leaving group of the transesterification reaction. (b) Active site of the hairpin ribozyme (Rupert et al., 2002). The cleavage reaction was inhibited by replacing the hydrogen atom of the nucleophile 2′-OH with a methyl group (magenta). The degree of impairment of the ribozyme when the catalytic nucleotides are replaced with abasic substitutions (Kuzmin et al., 2005; Lebruska et al., 2002) is indicated.