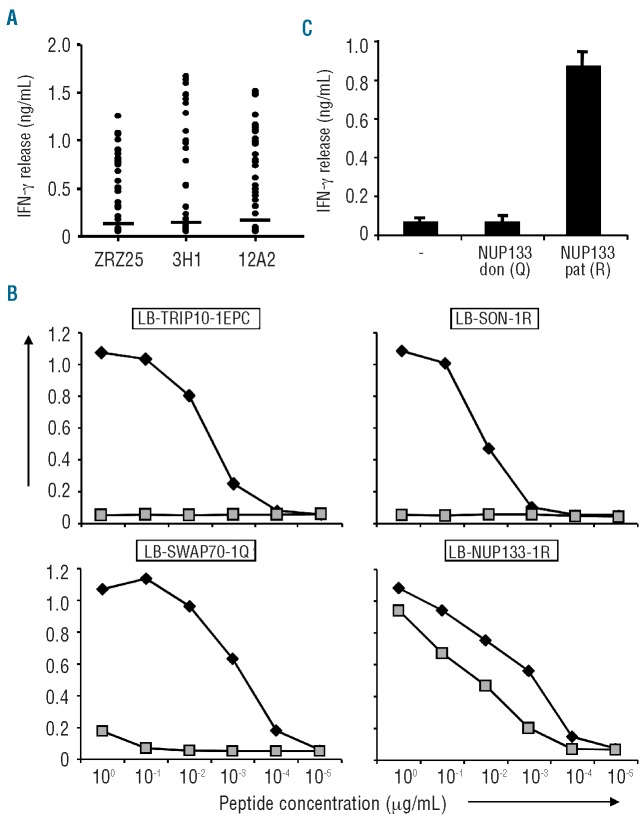
Figure 2.
Identification of MiHA by whole genome association scanning. (A) A panel of approx. 60 SNP-genotyped EBV-B cells expressing HLA-B*40:01 endogenously or after retroviral transfer of MP71-HLA-B*40:01-IRES-NGFR with more than 20% of marker gene positive cells were tested for recognition by T-cell clones ZRZ25, 3H1 and 12A2 in IFN-γ ELISA. EBV-B cells were divided into MiHA positive and negative groups based on a threshold of 5-fold the background production of IFN-γ (horizontal lines). Mean release of IFN-γ (ng/mL) in 50 μL culture supernatants of duplicate wells is shown. (B) Peptides comprising patient (filled symbols) type amino acids with predicted binding to HLA-B*40:01 as well as donor type peptide variants (gray symbols) were pulsed on donor EBV-B cells and tested for T-cell recognition in IFN-γ ELISA. LB-TRIP10-1EPC (GEPQDLCTL) contains 3 patient type amino acids and has been identified by cDNA library screening. LB-SON-1R (SETKQRTVL), LB-SWAP70-1Q (MEQLEQLEL) and LB-NUP133-1R (SEDLILCRL) contain single patient type amino acids encoded by exon SNPs identified by WGAs based on significant association with T-cell recognition. Donor type peptides SETKQCTVL and MEQLEELEL encoded by the genes for SON and SWAP70, respectively, were not recognized by the T-cell clones, whereas donor type peptide SEDLILCQL encoded by the NUP133 gene was similarly recognized as LB-NUP133-1R. Mean production of IFN-γ (ng/mL) in 50 μL culture supernatants of duplicate wells at various peptide concentrations (μg/mL) is shown. (C) NUP133 genes were isolated from patient and donor derived cDNA and cloned into expression vector pcDNA-3. Hela cells stably expressing HLA-B*40:01 were transiently transfected with patient and donor derived NUP133 genes encoding the R and Q at position 406, respectively, and incubated with T-cell clone 3H1. Mean production of IFN-γ (ng/mL) in 50 μL culture supernatants of duplicate wells is shown.