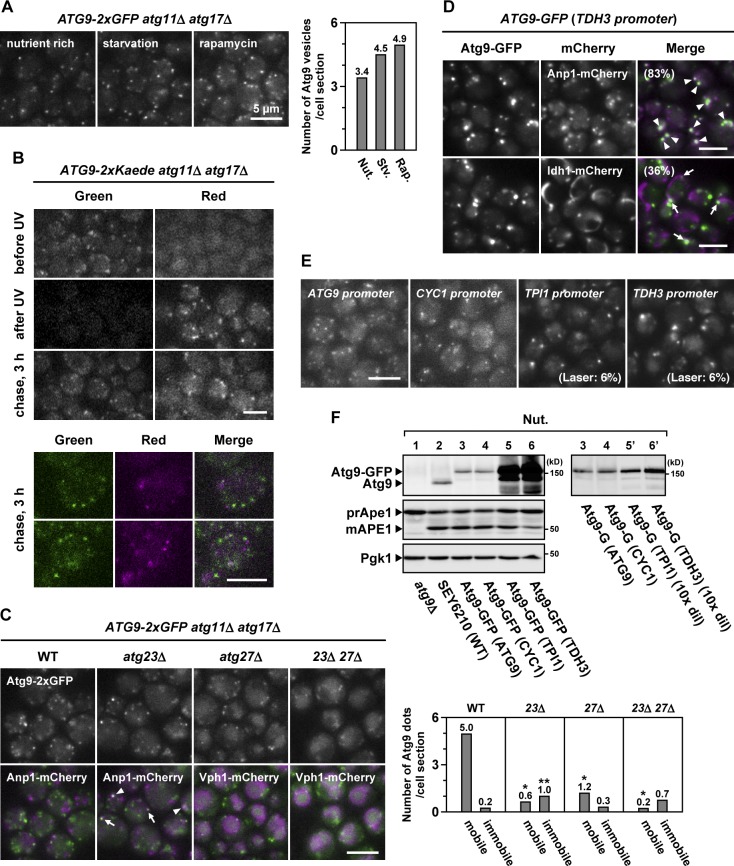
Figure 3.
Atg9 vesicles are generated de novo during starvation. (A) ATG9-2×GFP atg11Δ atg17Δ cells were either starved for 2 h or treated with rapamycin for 2 h. The number of Atg9 vesicles per cell section was counted. This experiment was completed once (n > 135). Nut., nutrient; Stv., starved; Rap., rapamycin. (B) ATG9-2×Kaede atg11Δ atg17Δ cells were treated with rapamycin for 1 h (before UV) and then irradiated with 365-nm UV (after UV). The cells were subjected to chase incubation for 3 h (chase, 3 h). (C) ATG9-2×GFP atg11Δ atg17Δ cells lacking Atg23 and/or Atg27 were treated with rapamycin for 3 h (see also Video 5). Anp1-mCherry (Golgi) and Vph1-mCherry (vacuole) were used as organelle markers. Arrowheads and arrows indicate Atg9-GFP clusters accumulated at and adjacent to the Golgi apparatus, respectively. The number of mobile Atg9 dots and immobile Atg9 dots per cell section was counted. This experiment was completed once (n > 250). Single asterisks indicate, in these mutant cells, very small mobile Atg9 dots that were observed but not counted. The double asterisk indicates, in atg23Δ cells, immobile Atg9 dots that were occasionally located at or adjacent to the Golgi apparatus (42%). (D) Cells expressing Atg9-GFP via the TDH3 promoter were observed at 32 ms/frame. Anp1-mCherry (Golgi) and Idh1-mCherry (mitochondria) were used as organelle markers. Arrowheads and arrows indicate immobile Atg9-GFP clusters adjacent to the Golgi apparatus and mitochondria, respectively. (E) Cells expressing Atg9-GFP via the ATG9 promoter, CYC1 promoter, TPI1 promoter, or TDH3 promoter were observed at 32 ms/frame (see also Video 6). When Atg9-GFP was expressed via the TPI1 promoter or TDH3 promoter, the excitation laser intensity was lowered to 6% with a neutral density filter. (F) Total cell lysates were prepared from the cells used in E and subjected to immunoblotting. Some samples were diluted 10-fold (10× dil). WT, wild type.