Abstract
Astrocyte elevated gene-1 (AEG-1), also known as MTDH and Lyric, is a novel gene that was first cloned by subtraction hybridization in 2002 and has recently been shown to play a role as a crucial oncogene that acts as a promoter of tumor malignancy. Overexpression and inhibition studies both in in vitro and in vivo models have partly shown the oncogenic roles of AEG-1 in a number of crucial aspects of tumor development and progression, including transformation, evasion of apoptosis, proliferation, cell survival, migration, invasion, metastasis, angiogenesis and chemoresistance through the activation of numerous signaling pathways, such as the nuclear factor κB, PI3K/AKT, Wnt/β-catenin and mitogen-activated protein kinase signaling pathways. However the potential roles of AEG-1, particularly in specific organs or tissues, such as breast tissue, require further clarification. Studies have found that in normal human breast tissue, AEG-1 is always expressed at low levels or is absent, while it is widely overexpressed in many breast cancer cell lines and breast tumors. The present review evaluates the current literature with regards to AEG-1 relative to breast cancer development and progression and highlights new perspectives relative to this molecule, indicating its potential to become a new target for the clinical treatment of breast cancer.
Keywords: astrocyte elevated gene-1, metastasis, angiogenesis, prognosis, breast cancer
1. Introduction
Breast cancer is the most common type of cancer in females worldwide. Statistics show that during the period between 2000 and 2005, the incidence of breast cancer in Europe was approximately 370,000 cases per year. This type of cancer is the most common cause of cancer-related death in women (1). Apart from the mortality caused by tumor degradation in situ, the majority of fatal cases are caused by local, regional or distant (bone, liver, lung, kidney, thyroid and brain) metastasis from the primary tumor site (breast tissues), which spreads through the bloodstream or lymphatic channels (2).
Tumorigenesis is a multistep process that transforms normal cells into the malignant phenotype, is always accompanied by genetic alterations. It frequently leads to the dysregulation of numerous signal transduction pathways, which are usually involved in cell cycle progression, cell death and survival, angiogenesis and apoptosis. Moreover, these pathways may present rational targets, providing opportunities for the targeted therapeutics of cancer (3). Advances in molecular technology have led to a better understanding of the mechanisms involved in the development and progression of breast cancer. Over the past few decades, an increasing body of knowledge regarding specific genes and proteins, such as HER2 and VEGF/VEGFR, as well as biological molecular pathways associated with the development and progression of breast cancer, has allowed for the development of targeted therapeutics that ideally aim at the selective, efficient and safe treatment of cancer. These developments have greatly improved the survival of breast cancer patients, while both chemotherapy and hormone therapy have also significantly prolonged the survival of breast cancer patients (3,4).
Following its initial cloning in 2002 (5), astrocyte elevated gene-1 (AEG-1) has been found to be overexpressed in multiple carcinomas, including melanoma, glioma and neuroblastoma, as well as carcinomas of the breast, prostate, liver and esophagus (6). In addition, AEG-1 has been found to play various roles in the regulation of cancer development, progression and metastasis.
Recent studies have shown that in normal human breast tissues, the expression of AEG-1 is decreased or is completely absent, while it is widely overexpressed in a number of breast cancer cell lines and breast tumors (7–10). Current investigations have revealed that AEG-1 plays a crucial role in the regulation of specific development and progression of breast cancer, indicating its potential as a specific new target in clinical-targeted therapeutics of breast cancer.
2. Cloning and molecular structure of AEG-1
Following human immunodeficiency virus (HIV)-1 infection or treatment with HIV-1 envelope glycoprotein (gp120) of primary human fetal astrocytes (PHFA), AEG-1 was found to be overexpressed by the rapid subtraction hybridization (RaSH) method (5). In 2003, a customized microarray approach used to detect the expression of AEG-1 in PHFA treated with tumor necrosis factor (TNF)-α showed a concomitant change similar to that following HIV-1 infection (11).
The AEG-1 gene consists of 12 exons and 11 introns and is located at 8q22 via genomic blast search. Excluding the poly-A tail, the full-length AEG-1 cDNA consists of 3,611 bp (8). The open reading frame (ORF) from 220 to 1,968 nt of human AEG-1 encodes a 582-amino acid protein with a calculated molecular mass of 64 kDa and a pI of 9.33. Protein motif analysis of the AEG-1 protein and three independent transmembrane protein prediction methods (PSORT II, TMpred and HMMTOP) predicted that the AEG-1 protein contains a single-transmembrane domain. Notably, PSORT II and TMpred predicted AEG-1 as a type Ib protein (C terminal in the cytoplasmic side with no signal peptide), whereas TMHMM and TopPred 2 obtained a reverse localization of the C terminal, indicating that it was a type II protein. Although the analysis of the amino acid sequence of AEG-1 revealed that it does not have any recognizable protein domains, the presence of putative, either monopartite or bipartite, nuclear localization signals (NLS) between amino acids 432–451 and 561–580 was confirmed, suggesting that it may enter into the nucleus to play a significant role (12). In another study (13), it was shown that AEG-1 contains three NLS which may regulate its distribution and function in cells. The final results obtained suggest that extended NLS-1 (amino acids 78–130) regulates its nucleolar localization, and the extended NLS-2 region (amino acids 415–486) mediates its modification via ubiquitination almost exclusively within the cytoplasm, whereas the COOH-terminal extended NLS-3 (amino acids 546–582) is the predominant regulator of nuclear localization (13). Moreover, AEG-1 was found to be rich in both lysine residues, which are targets for post-translational modification by ubiquitination, and serine residues, required for the redistribution of AEG-1 within the cell (13).
3. Intracellular localization of AEG-1
Intracellular localization of AEG-1 has been examined in various cell types, using a polyclonal anti-AEG-1 antibody and immunofluorescence microscopy. In immortalized PHFA (IM-PHFA), endogenous AEG-1 was stained. The image showed that AEG-1 was located in the perinuclear region and in endoplasmic reticulum (ER)-like structures, but not in the plasma membrane. It was colocalized with the ER-specific protein calreticulin, but not with the mitochondrial marker MitoTracker (8). Concomitantly, the observed ER localization of AEG-1 indicates that it is a type Ib membrane protein (8). However, following TNF-α treatment or forced overexpression of AEG-1, localization of the protein was detected both in the cytoplasm and nucleus in HeLa cells (12). This phenomenon is similar to that of the nuclear receptor coactivator SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1), which is located mainly in the cytoplasm and translocates from the cytoplasm to the nucleus in response to TNF-α (14), indicating a potential direct or indirect interaction between AEG-1 and SRC-3 or other unknown factors. Although various studies (8,12) have been performed to investigate the intracellular localization of AEG-1, the specific localization and the relationship between its localization and function require further clarification.
4. Molecular mechanism of AEG-1 action
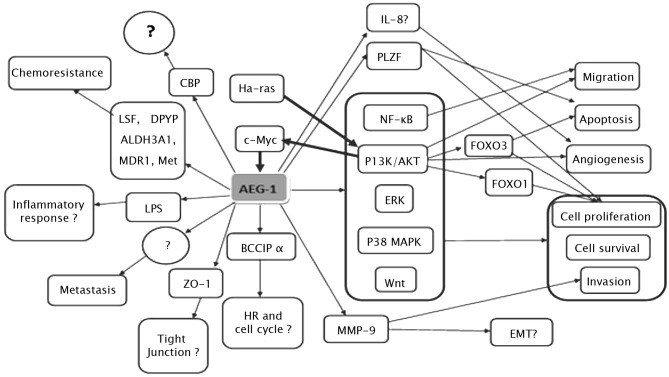
AEG-1 contributes to several hallmarks of metastatic cancers, including proliferation, evasion of apoptosis and cell survival under stressful conditions, such as serum deprivation and chemotherapy. AEG-1 also enhances migration, invasion, angiogenesis and metastasis by modulating various signaling pathways (Fig. 1). It is well established that the Ha-ras and c-Myc genes cooperate to promote transformation, tumor development and progression, and metastasis. However, the precise mechanism underlying this cooperation remains to be determined. The overexpression of AEG-1 and oncogenic Ha-Ras has been found to collaborate to enhance the soft agar colony formation of immortalized melanocytes and astrocytes (8). It was observed that human AEG-1 can be induced at the transcription level by oncogenic Ras in human fetal astrocytes, whereas AEG-1 knockdown suppressed Ras-induced colony formation (15). Moreover, when inhibitors for various Ras downstream signaling pathways were examined, only PI3K/AKT inhibitors LY294002 and PTEN were able to block AEG-1 promoter activation by Ras, suggesting the involvement of the PI3K/AKT pathway in Ha-ras-induced AEG-1 regulation (15). To confirm that the PI3K/AKT signaling pathway is involved in oncogenic Ha-ras-induced AEG-1 expression, the phosphorylation of AKT and GSK3β was analyzed by Western blotting, indicating a parallel overexpression of AKT and GSK3β. Promoter mapping subsequently identified the region −356 to −302 of the AEG-1 promoter containing two E-box elements that bind to Ha-ras-activated transcription factors, such as c-Myc.
Figure 1.
Hypothetical model of the signal transduction pathways associated with the development and progression of cancer, which are regulated by AEG-1 (thin arrows) and the signal transduction pathways modulating the expression of AEG-1 (thick arrows). The contents marked by symbol ‘?’ indicate the issues requiring further elucidation.
All of the above data indicate that Ha-ras increases binding of c-Myc to the E-box elements in the AEG-1 promoter through the PI3K/AKT/GSK3β/c-Myc pathway to act in Ha-ras-mediated oncogenesis through AEG-1 (15). However, inhibition of the MEK pathway was found to slightly increase Ha-ras-mediated AEG-1 promoter activation, the significance of which remains to be determined (15). Notably, transcription of AEG-1 is induced by c-Myc, whereas AEG-1 induces c-Myc expression (in neuroblastoma cells it induces N-Myc expression), thereby amplifying the tumorigenic effect (16). A recent study showed that one mechanism by which cells with altered AEG-1 expression evade apoptosis and increase cell growth during tumorigenesis involves the repression of the function of promyelocytic leukemia zinc finger (PLZF) (17). The transcriptional repressor PLZF, which has been found to regulate the expression of genes involved in cell growth and apoptosis, including c-Myc, was found to interact with AEG-1 through a yeast two-hybrid screening system. It was also noted that AEG-1 interacts with PLZF, reducing PLZF binding to promoters, thereyb reducing PLZF-mediated repression (17).
AEG-1 activates several downstream signal transduction pathways, including the nuclear factor κB (NF-κB), PI3K/AKT, Wnt/β-catenin and MAPK pathways to enhance various aspects of tumor development and progression. The first signaling pathway identified as being activated by AEG-1 was NF-κB (12). In HeLa cells, following infection with Ad.AEG-1 or treatment with TNF-α, AEG-1 was found to translocate into the nucleus where it interacted with the p65 subunit of NF-κB and enhanced NF-κB-induced gene expression. AEG-1 activates NF-κB via IκBα degradation and p65 translocation (12). A gene array analysis revealed that Ad.AEG-1 infection resulted in a marked up-regulation of NF-κB-responsive cell adhesion molecules (ICAM-2 and ICAM-3, selectin E, selectin L and selectin P ligand), TLR4 and TLR5, and cytokines, such as IL-8 (12). Following AEG-1 overexpression or TNF-α treatment, AEG-1 was found to interact with p65 and cyclic AMP-responsive element binding protein-binding protein (CBP) on the IL-8 promoter, increasing IL-8 transcription, suggesting that AEG-1 acts as a bridging factor among NF-κB, CBP and the basal transcription mechanisms (18). AEG-1-induced increase of soft agar growth and matrigel invasion may be restrained by the inhibition of NF-κB in HeLa cells (12).
The present review elucidates the domains of AEG-1 that are crucial for mediating its role. Functional analysis revealed that the functions of AEG-1 in invasion, migration and NF-κB-activating properties are mediated by NH2-terminal 71 amino acids, whereas amino acids 101 to 205 were identified as a p65-interaction domain, indicating that p65 interaction alone is not sufficient to mediate AEG-1 function (18). NF-κB activation by AEG-1 has also recently been documented in prostate and liver cancer cells (19,20). Notably, AEG-1 was found to be induced via NF-κB activation in LPS-stimulated U937 human promonocytic cells, and AEG-1 induced by LPS subsequently regulated NF-κB activation in turn. Moreover, LPS-induced TNF-α and prostaglandin E2 production were inhibited by preventing the expression of AEG-1. Therefore, we hypothesize that AEG-1 is a LPS-responsive gene and is involved in LPS-induced inflammatory response (21).
PI3K/AKT is another pathway involved in the tumorigenesis mediated by AEG-1. This pathway is not only activated by AEG-1, but also regulates the expression of AEG-1 (15). PI3K/AKT signals modulates various growth-regulatory transcription factors, such as the forkhead box (FOXO) protein and NF-κB. AEG-1 knockdown was found to induce cell apoptosis through the up-regulation of FOXO3a activity in prostate cancer cells, and subsequently revealed that AEG-1 expression plays a dominant role as a positive auto-feedback activator of AKT and as a suppressor of FOXO3a to promote PC progression (20). Moreover, it was noted that AEG-1 knockdown down-regulated the constitutive activity of NF-κB and the activator protein 1 (AP-1), while the mRNA and protein expression levels of NF-κB and AP-1-regulated genes IL-6, IL-8 and matrix metalloproteinase-9 (MMP-9) were significantly decreased. The invasive properties of PC-3 and DU145 cells were also found to be decreased (20).
AEG-1 provides protection from serum starvation-induced apoptosis of normal cells by activating the PI3K/AKT pathway (22). In order to clarify the molecular mechanisms underlying AEG-1-dependent enhanced cell growth under conditions of serum starvation, the downstream antiapoptotic substrates of AKT were examined, revealing that the phosphorylation-induced inactivation of GSK3β increased c-Myc levels, inhibited serum starvation-dependent p21/mda-6 expression and phosphorylation of Bad, a proapoptotic member of the Bcl-2 family, in Ad.AEG-1-infected PHFA (22). In less aggressive neuroblastoma cells, the overexpression of AEG-1 enhanced proliferation and expression of the transformed state by activating the PI3KAKT signaling pathway (16). In esophageal cancer cells, the up-regulation of AEG-1 reduced the expression of p27 and induced the expression of cyclin D1 through the AKT/FOXO3a pathway (23).
The PI3K/AKT pathway regulates AEG-1-induced angiogenesis (24). An immunohistochemical analysis revealed that tube formation induced by enhanced AEG-1 expression correlates with an increased expression of specific angiogenesis molecules, including angiopoietin-1 (Ang1), MMP-2 and hypoxia-inducible factor 1-α (HIF1-α) in tumor sections. To analyze the potential importance of the PI3K/AKT signaling pathway in AEG-1-mediated angiogenesis, Ad.DN.AKT was constructed. The result showed that AEG-1-mediated endothelial cell tube formation was significantly inhibited by Ad.DN.AKT, suggesting that the PI3K/AKT pathway is an integral component of this process (24).
AEG-1 has also been associated with the Wnt/β-catenin pathway in hepatocellular carcinoma (HCC) cells through the activation of the Raf/MEK/MAPK branch of the Ras signaling pathway (19). In HCC, AEG-1 was found to activate Wnt/β-catenin signaling by activating ERK42/44 and up-regulating lymphoid-enhancing factor 1/T-cell factor 1 (LEF1/TCF1), the ultimate executor of the Wnt pathway (19). Moreover, specific inhibitors of the MAPK pathway are able to abolish the oncogenic effect of AEG-1 on Matrigel invasion and anchorage-independent growth, indicating that AEG-1 also plays an oncogenic role in tumor development and progression through activation of the MAPK signal pathway (19).
5. AEG-1 and proliferation of breast cancer
Uncontrolled cell growth is largely associated with alterations in genes or proteins related to the regulation of proliferation, cell death, apoptosis and genetic stability, such as tumor suppressor genes, oncogenes, growth factors and cell adhesion molecules, which vary among different cancer types (25). Findings of a recent study showed that AEG-1 promotes proliferation of breast cancer cells by down-regulating the transcriptional activity of FOXO1 by inducing its phosphorylation through the PI3K/AKT signaling pathway (26). Up-regulation of AEG-1 was found to markedly promote proliferation (detected by MTT) and tumorigenicity (tested by anchorage-independent growth) in MCF-7 and MDA-MB-435 breast cancer cells, whereas an AEG-1-knockdown cell model with shRNAs inhibited cell proliferation and the colony-forming ability of cells in soft agar. Furthermore, since these proliferative effects were significantly associated with decreases in p27Kip1 and p21Cip1, two key cell cycle inhibitors, overexpression of AEG-1 in breast cancer cells may significantly impact cell cycle checkpoints, thereby promoting the proliferation of breast cancer cells (26). In addition, BCCIP α expression was reduced in human breast tissue and it was found that when the BCCIP α protein was re-introduced into MCF7 breast cancer cells, the growth of breast cancer cells was inhibited (27). Studies have shown the multifaceted roles played by BCCIP α. It binds to p21 and enhances the p21-mediated inhibition of Cdk2 kinase, whereas loss of BCCIP impairs G1/S checkpoint activation following DNA damage. BCCIP also plays a role in homologous recombination repair of DNA damage by interacting with BRCA2 and contributes to the maintenance of chromosome stability (27–30). Recently, it was found that AEG-1 promotes proteasomal degradation of tumor suppressor BCCIP α (29). Therefore, whether AEG-1 promotes proliferation of breast cancer through the regulation of the expression of BCCIP α should be confirmed. Moreover, AEG-1 was found to promote the proliferation of various types of cancer cells by activating the PI3K/AKT and NF-κB signaling pathways. Thus, apart from the PI3K/AKT signaling pathway, whether the ectopic expression of AEG-1 enhances the proliferation and anchorage-independent growth of breast cancer cells by activating other prosurvival signaling pathways, such as NF-κB, remains to be determined.
6. AEG-1 and the invasion and migration of breast cancer
Overexpression of AEG-1 significantly enhances the invasive ability of cells as revealed by Matrigel invasion assay in HeLa cells, while inhibition of AEG-1-induced NF-κB activation blocks invasion (12). Overexpression of AEG-1 increases, whereas siRNA inhibition of AEG-1 decreases the migration and invasion of human glioma cells, respectively (31). AEG-1 was also found to induce invasion in normal immortal cloned rat embryo fibroblast (CREF) cells (24). During the early progression of metastasis, a number of signal transduction pathways are activated, including PI3K/AKT and NF-κB. The two signaling pathways have been found to be linked to the promotion of cancer cell invasion and migration through the down-regulation of cell-cell contact protein E-cadherin and the up-regulation of MMP-2 and MMP-9 (32–34). It is known that AEG-1 activates the PI3K/AKT and NF-κB signaling pathways, and AEG-1 has been found to up-regulate MMP-9 and induce human glioma cell invasion (35). Although experiments investigating the invasion and migration of breast cancer have yet to be conducted, it is thought that AEG-1 promotes the invasion and migration of breast cancer by activating the PI3K/AKT and/or NF-κB signaling pathways, or other signaling pathways, which requires confirmation.
7. AEG-1 and metastasis of breast cancer
Tumor metastasis is a complex multistep process in which cancer cells detach from the primary tumor tissue and establish metastatic foci at specific sites. Brown and Ruoslahti (9)used a phage expression library of cDNAs from metastatic breast carcinoma to identify protein domains that bind to lung vasculature. These authors found an extracellular lung-homing domain (LHD; amino acids 378–440 in mouse or 381–443 in human) in AEG-1 to be a mediator of 4T1 mouse mammary tumor cell adhesion to lung vasculature (9). In addition, antibodies to AEG-1 showed a high expression of AEG-1 throughout human breast tumors and breast tumor xenografts, while markedly lower levels of AEG-1 were present in normal breast tissue (9). Moreover, antibodies reactive to the lung-homing domain of AEG-1 and siRNA-mediated knockdown of AEG-1 expression inhibited experimental breast cancer lung metastasis. Conversely, enhanced localization to lung vasculature was noted in HEK293T cells transiently transfected with AEG-1. These results suggest that AEG-1 plays an important role in breast cancer metastasis (9). To further clarify the mechanism of metastasis of breast cancer, human breast cancer cell line MDA-MB-231 was used to investigate the roles of a number of genes in metastasis, including UQCRB, PTDSS1, TSPYL5, AEG-1, LAPTM4β and SDC2 (10). The data showed that AEG-1 overexpression significantly accelerated the development of lung metastasis and led to a modest, but significant, increase in bone and brain metastases (10). These results suggest that AEG-1 preferentially promotes metastasis to the lung, but has a modest impact on metastasis to other organs. As mentioned above, the PI3K/AKT and NF-κB signaling pathways are involved in the early progression of metastasis. AEG-1 may promote metastasis through the interaction of the LHD, with an unknown receptor expressed in the surface of endothelial cells, or indirectly through the activation of signaling pathways, such as PI3K/AKT and NF-κB, which involve adhesion molecules, such as MMPs.
8. AEG-1 and angiogenesis of breast cancer
Hanahan and Weinberg proposed that six essential ‘hallmarks’ or processes are required for the transformation of a normal cell to a cancer cell. These processes include: i) self-sufficiency in growth signals; ii) insensitivity to antigrowth signals; iii) evasion of programmed death (apoptosis); iv) endless replication potential; v) sustained angiogenesis; and vi) tissue invasion and metastasis (36). Overexpression of AEG-1 increases the expression of molecular markers of angiogenesis, including Ang1, MMP-2 and HIF1-α (24). In vitro angiogenesis studies further demonstrated that AEG-1 promotes tube formation in Matrigel and increases invasion of human umbilical vein endothelial cells via the PI3K/AKT signaling pathway (24). The angiogenesis-promoting function of AEG-1 was also noted in human HCC (19). Numerous genes play important roles in angiogenesis, apart from VEGF and VEGFR, which are the most representative. IL-8 and COX2 also promote tumor angiogenesis, but are not involved in the VEGF pathway (37–39). Many genes are involved in the NF-κB signaling pathway, such as IL-8 and COX2. IL-8 and COX2 are overexpressed in breast cancer (40,41), and the expression of IL-8 can be regulated by AEG-1 (12), indicating that AEG-1 may also promote the angiogenesis of breast cancer by activating the PI3K/AKT signaling pathway as well as the NF-κB or VEGF signaling pathway. Experiments in vivo and in vitro should be performed to clarify the mechanism.
9. AEG-1 and chemoresistance of breast cancer
AEG-1 plays a role in chemoresistance in human HCC, neuroblastoma cell lines and breast cancer (10,19,40). An Affymetrix oligonucleotide microarray was performed in human HCC to analyze the downstream genes of AEG-1. The results of the microarray showed a cluster of genes associated with chemoresistance, including drug-metabolizing enzymes for various chemotherapeutic agents, such as dihydropyrimidine dehydrogenase (DPYD), principal enzyme-inactivating 5-fluorouracil (5-FU) cytochrome P4502B6 (CYP2B6), dihydrodiol dehydrogenase (AKR1C2) and the ATP-binding cassette transporter ABCC11 for drug efflux (19). AEG-1 was also found to enhance the expression of the transcription factor LSF, which regulates the expression of thymidylate synthase, a target of 5-FU. In addition, AEG-1 enhanced the expression of dihydropyrimidine dehydrogenase (DPYD), which catalyzes the initial and rate-limiting step in the catabolism of 5-FU (42). A recent study reported a new mechanism through which AEG-1 plays a role in chemoresistance in human HCC (43). AEG-1 increases the expression of multidrug resistance gene 1 (MDR1) protein, resulting in increased efflux and decreased accumulation of doxorubicin, thereby promoting doxorubicin resistance (43). AEG-1 increases the expression of MDR1 by facilitating the association of MDR1 mRNA to polysomes, resulting in increased translation. AEG-1 also inhibits ubiquitination and subsequent proteasome-mediated degradation of the MDR1 protein (43). In neuroblastoma cells, a significant enhancement in chemosensitivity to cisplatin and doxorubicin by knockdown of AEG-1 was observed (40). In breast cancer, in vitro and in vivo analyses of chemoresistance revealed that AEG-1 knockdown sensitized various breast cancer cell lines (LM2, MDA-MB-231, MCF7 and T47D) to paclitaxel, doxorubicin, cisplatin, 4-hydroxycylcophosphamide and hydrogen peroxide (10). AEG-1 did not affect the uptake or retention of chemotherapy drugs in breast cancer tissue (10). Instead, AEG-1 may increase chemoresistance by promoting cellular survival against antineoplastic stresses (10).
Since we confirmed that AEG-1 activates the PI3K/AKT and NF-κB signaling pathways, chemoresistance may be mediated by these prosurvival pathways through the interaction between AEG-1 and molecules involved in these pathways. Microarray analysis of AEG-1-knockdown breast cancer cells revealed that two AEG-1-down-regulated genes (TRAIL and BINP3, two cell death-inducing genes) and a number of AEG-1-up-regulated genes (ALDH3A1, MET, HSP90 and HMOX1) are involved in chemoresistance (10). Among these candidate AEG-1 downstream genes, ALDH3A1 (aldehyde dehydrogenase 3 family, member A1) and MET (hepatocyte growth factor receptor) collectively contribute to its role in broad-spectrum chemoresistance (10). Whether AEG-1 regulates MDR1 expression in breast cancer and whether AEG-1 increases chemoresistance by promoting cellular survival or regulating downstream genes that directly modulate chemoresistance require elucidation. Moreover, the specific mechanism of chemoresistance of AEG-1 in breast cancers remains to be clarified.
10. AEG-1 and prognosis of breast cancer
Two independent research groups analyzed AEG-1 expression and clinical associations, respectively, using breast tumor samples and obtained similar results (7,10). AEG-1 was found to be abundantly expressed in breast tumors with a percentage of 47 and 44.4%, respectively, in these two groups, and was significantly correlated with clinical staging, tumor size, lymph node spread, distant metastasis and poor survival (7,10). Overexpression of AEG-1 was not linked to any specific breast tumor subtype in terms of HER2 status, triple marker status (ER/PR/HER2), or basal epithelial cell marker CK5/6 status (10). Findings of the multivariate analysis suggested that AEG-1 expression may be an independent prognostic indicator independent of other clinicopathological factors for the survival of patients with breast cancer (7,10). Further evidence points to the fact that AEG-1 is located at chromosome 8q22, a region frequently amplified in many cancer types and associated with poor prognosis (44–47). These findings suggest the clinical and prognostic significance of AEG-1 in breast cancer.
11. Conclusion and future perspectives
Although in vitro and in vivo studies have confirmed that AEG-1 plays a key role in the process of cancer development and progression in multiple organs, including breast, further investigations should be conducted to clarify the specific mechanisms involved in the mediation of AEG-1 functions. Moreover, additional unknown functions of AEG-1 should be further studied (48).
Since we know that a correlation exists between AEG-1 and tumor progression, whether AEG-1 regulates other functions related to tumor progression, such as immortalization and transformation, as suggested by numerous studies in other types of cancers, and whether using immortal normal cells in breast cancer is viable should be investigated. For example, local invasion is considered to be a crucial first step in metastatic dissemination, and epithelial-mesenchymal transition (EMT) and epithelial plasticity are hypothesized to contribute to tumor progression (49). In breast cancer, features of EMT have been observed (50) and it was found that developmental EMT regulators, including Snail/Slug, Twist, Six1 and Cripto, along with developmental signaling pathways, including TGF-β and Wnt/β-catenin, are aberrantly expressed in breast cancer (49). MMPs also induce EMT in breast cancer (51). Since we found that AEG-1 regulates the expression of MMP-9 and activates the Wnt/β-catenin signaling pathway, whether AEG-1 is involved in the regulation of EMT in breast cancer through the modulation of regulators, such as MMP-9 or through the activation of signaling pathways, such as Wnt/β-catenin, requires clarification. In addition, data show that AEG-1 colocalizes with tight junction proteins ZO-1 and occludin in polarized epithelial cells (52), suggesting that AEG-1 is related to the loss of cell polarity known to occur with increased epithelial tumorigenicity. Previous findings have shown that tight junctions are comprised of a number of types of membrane proteins, cytoskeletal proteins and signaling molecules, many of which play roles in the development of the mammary gland. Moreover, several of these proteins are regarded as potential ‘tumor suppressors’ during the development and progression of breast cancer (53). EMT is associated with the disintegration of tight junctions (54). These findings indicate potential new roles of AEG-1 in tight junctions and EMT.
Acknowledgements
This study was partly supported by the Program for New Century Excellent Talents in University, Key Project of Chinese Ministry of Education (no. 108080), the National Natural Science Foundation of China (nos. 30772133 and 81072150) and the Independent Innovation Foundation of Shandong University (IIFSDU) (no. 2009JQ007) to Professor Q. Yang.
References
- 1.Krcova Z, Ehrmann J, Krejci V, Eliopoulos A, Kolar Z. Tpl-2/Cot and COX-2 in breast cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2008;152:21–25. doi: 10.5507/bp.2008.003. [DOI] [PubMed] [Google Scholar]
- 2.Jezierska A, Motyl T. Matrix metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:RA32–RA40. [PubMed] [Google Scholar]
- 3.Gasparini G. Therapy of breast cancer with molecular targeting agents. Ann Oncol. 2005;16:iv28–iv36. doi: 10.1093/annonc/mdi905. [DOI] [PubMed] [Google Scholar]
- 4.Osborne C, Wilson P, Tripathy D. Oncogenes and tumor suppressor genes in breast cancer: potential diagnostic and therapeutic applications. Oncologist. 2004;9:361–377. doi: 10.1634/theoncologist.9-4-361. [DOI] [PubMed] [Google Scholar]
- 5.Su ZZ, Kang DC, Chen YM, et al. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- 6.Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15:5615–5620. doi: 10.1158/1078-0432.CCR-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Zhang N, Song LB, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 8.Kang D, Su Z, Sarkar D, Emdad L, Volsky D, Fisher P. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 10.Hu G, Chong R, Yang Q, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su Z. Customized rapid subtraction hybridization (RaSH) gene microarrays identify overlapping expression changes in human fetal astrocytes resulting from human immunodeficiency virus-1 infection or tumor necrosis factor-α treatment. Gene. 2003;306:67–78. doi: 10.1016/s0378-1119(03)00404-9. [DOI] [PubMed] [Google Scholar]
- 12.Emdad L. Activation of the nuclear factor κB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 13.Thirkettle HJ, Girling J, Warren AY, et al. LYRIC/AEG-1 is targeted to different subcellular compartments by ubiquitinylation and intrinsic nuclear localization signals. Clin Cancer Res. 2009;15:3003–3013. doi: 10.1158/1078-0432.CCR-08-2046. [DOI] [PubMed] [Google Scholar]
- 14.Wu RC, Qin J, Hashimoto Y, et al. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by IκB kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci USA. 2006;103:17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SG, Jeon HY, Su ZZ, et al. Astrocyte elevated gene-1 contributes to the pathogenesis of neuroblastoma. Oncogene. 2009;28:2476–2484. doi: 10.1038/onc.2009.93. [DOI] [PubMed] [Google Scholar]
- 17.Thirkettle HJ, Mills IG, Whitaker HC, Neal DE. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene. 2009;28:3663–3670. doi: 10.1038/onc.2009.223. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-κB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 19.Yoo BK, Emdad L, Su ZZ, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikuno N, Shiina H, Urakami S, et al. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26:7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- 21.Khuda II, Koide N, Noman AS, et al. Astrocyte elevated gene-1 (AEG-1) is induced by lipopolysaccharide as toll-like receptor 4 (TLR4) ligand and regulates TLR4 signalling. Immunology. 2009;128:e700–e706. doi: 10.1111/j.1365-2567.2009.03063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF, Fisher PB. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene. 2007;27:1114–1121. doi: 10.1038/sj.onc.1210713. [DOI] [PubMed] [Google Scholar]
- 23.Yu C, Chen K, Zheng H, et al. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. doi: 10.1093/carcin/bgp064. [DOI] [PubMed] [Google Scholar]
- 24.Emdad L, Lee SG, Su ZZ, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci USA. 2009;106:21300–21305. doi: 10.1073/pnas.0910936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doerfler W, Hohlwey U, Müller K, Remus R, Heller H, Hertz J. Foreign DNA integration perturbations of the genome oncogenesis. Ann NY Acad Sci. 2001;945:276–288. doi: 10.1111/j.1749-6632.2001.tb03896.x. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Yang L, Song L, et al. Astrocyte elevated gene-1 is a proliferation promoter in breast cancer via suppressing transcriptional factor FOXO1. Oncogene. 2009;28:3188–3196. doi: 10.1038/onc.2009.171. [DOI] [PubMed] [Google Scholar]
- 27.Liu JM, Yuan Y, Huan J, Shen ZY. Inhibition of breast and brain cancer cell growth by BCCIPalpha, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene. 2001;20:336–345. doi: 10.1038/sj.onc.1204098. [DOI] [PubMed] [Google Scholar]
- 28.Meng XB, Lu HM, Shen ZY. BCCIP functions through p53 to regulate the expression of p21Waf1/Cip1. Cell Cycle. 2004;3:1457–1462. doi: 10.4161/cc.3.11.1213. [DOI] [PubMed] [Google Scholar]
- 29.Ash S, Yang D, Britt D. LYRIC/AEG-1 overexpression modulates BCCIPα protein levels in prostate tumor cells. Biochem Biophys Res Commun. 2008;371:333–338. doi: 10.1016/j.bbrc.2008.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng X, Fan J, Shen Z. Roles of BCCIP in chromosome stability and cytokinesis. Oncogene. 2007;26:6253–6260. doi: 10.1038/sj.onc.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emdad L, Sarkar D, Su Z, et al. Astrocyte elevated gene-1: recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther. 2007;114:155–170. doi: 10.1016/j.pharmthera.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunnumakkara A, Anand P, Aggarwal B. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Qiao M, Sheng SJ, Pardee AB. Metastasis and Akt activation. Cell Cycle. 2008;7:2991–2996. doi: 10.4161/cc.7.19.6784. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y. Opinion: emerging mechanisms of tumour lymphangiogenesis and lymphatic metastasis. Nat Rev Cancer. 2005;5:735–743. doi: 10.1038/nrc1693. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Wu J, Ying Z, et al. Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res. 2010;70:3750–3759. doi: 10.1158/0008-5472.CAN-09-3838. [DOI] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 37.Matsuo Y, Ochi N, Sawai H, et al. CXCL8/IL-8 and CXCL12/SDF-1α co-operatively promote invasiveness and angiogenesis in pancreatic cancer. Int J Cancer. 2009;124:853–861. doi: 10.1002/ijc.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuwano T. Cyclooxygenase 2 is a key enzyme for inflammatory cytokine-induced angiogenesis. FASEB J. 2004;18:300–310. doi: 10.1096/fj.03-0473com. [DOI] [PubMed] [Google Scholar]
- 39.Marrogi A, Travis W, Welsh J, et al. Nitric oxide synthase, cyclooxygenase 2, and vascular endothelial growth factor in the angiogenesis of non-small cell lung carcinoma. Clin Cancer Res. 2000;6:4739–4744. [PubMed] [Google Scholar]
- 40.Liu H, Song X, Liu C, Xie L, Wei L, Sun R. Knockdown of astrocyte elevated gene-1 inhibits proliferation and enhancing chemo-sensitivity to cisplatin or doxorubicin in neuroblastoma cells. J Exp Clin Cancer Res. 2009;28:19. doi: 10.1186/1756-9966-28-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerebours F, Vacher S, Andrieu C, et al. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo BK, Gredler R, Vozhilla N, et al. Identification of genes conferring resistance to 5-fluorouracil. Proc Natl Acad Sci USA. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo BK, Chen D, Su ZZ, et al. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 2010;70:3249–3258. doi: 10.1158/0008-5472.CAN-09-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DH, Mohapatra G, Bollen A, Waldman FM, Feuerstein BG. Chromosomal abnormalities in glioblastoma multiforme tumors and glioma cell lines detected by comparative genomic hybridization. Int J Cancer. 1995;60:812–819. doi: 10.1002/ijc.2910600615. [DOI] [PubMed] [Google Scholar]
- 45.Ried TJK, Holtgreve-Grez H, et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415–5423. [PubMed] [Google Scholar]
- 46.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 47.Warr T, Ward S, Burrows J, et al. Identification of extensive genomic loss and gain by comparative genomic hybridisation in malignant astrocytoma in children and young adults. Genes Chromosomes Cancer. 2001;31:15–22. doi: 10.1002/gcc.1113. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar D, Emdad L, Lee SG, Yoo BK, Su ZZ, Fisher PB. Astrocyte elevated gene-1: far more than just a gene regulated in astrocytes. Cancer Res. 2009;69:8529–8535. doi: 10.1158/0008-5472.CAN-09-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trimboli AJ, Fukino K, de Bruin A, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 51.Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britt D, Yang D, Flanagan D, et al. Identification of a novel protein, LYRIC, localized to tight junctions of polarized epithelial cells. Exp Cell Res. 2004;300:134–148. doi: 10.1016/j.yexcr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 53.Itoh M, Bissell MJ. The organization of tight junctions in epithelia: implications for mammary gland biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:449–462. doi: 10.1023/B:JOMG.0000017431.45314.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikenouchi J. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]