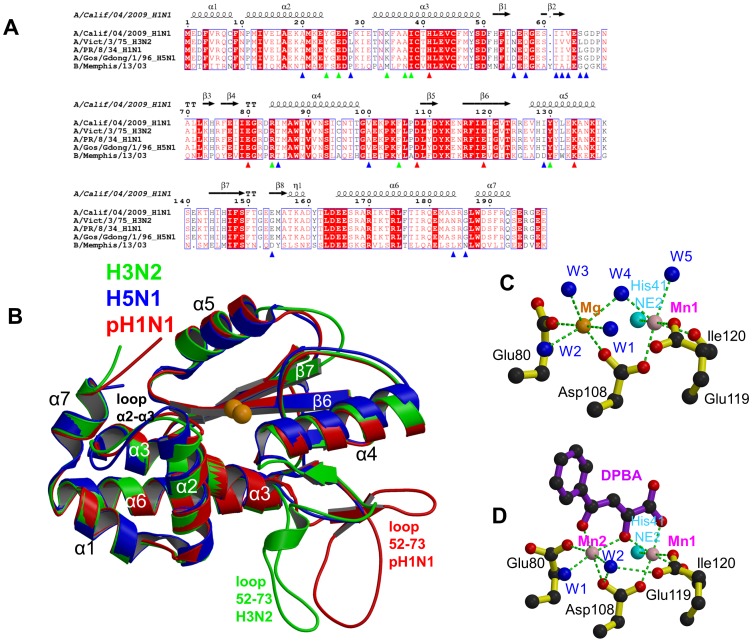
Figure 1. The PA endonuclease carries a divalent cation binding site in its active center.
A: Sequence alignment of the PA-Nter endonuclease from four influenza A (including the three of known atomic structure) and one influenza B strain. The secondary structure of the pH1N1 domain is shown over the alignment. Red triangles indicate conserved cation binding (His41, Glu80, Asp108, Glu119) and catalytic (Lys134) residues. Blue triangles indicate naturally variable positions amongst influenza A strains. Green triangles indicate residues interacting with the inhibitors described in this paper. B: Superposition of PA endonuclease structure from H3N2 (green, PDB entry 2W69), H5N1 (blue, PDB entry 3EBJ) and pH1N1 (red, this work). The two bound divalent metal ions are represented by orange spheres. Flexible region 53–73 is at the bottom right and only ordered in certain chains from the H3N2 (B chain) and pH1N1 (e.g. D chain of dTMP complex) structures. For H5N1, region 53–73 is not visible. Major secondary structure elements are shown consistent with those in Figure 1A. C: Divalent ion co-ordination in the native endonuclease structure. Manganese and magnesium ions are respectively pink and orange spheres and co-ordinating water molecule blue spheres and the ion co-ordination is shown with green dotted lines. For clarity, only His41 NE2 is shown (cyan sphere). D: Divalent ion co-ordination in the DPBA bound structure. Manganese ions are pink spheres and co-ordinating water molecule blue spheres and the ion co-ordination is shown with green dotted lines. For clarity, only His41 NE2 is shown (cyan sphere).