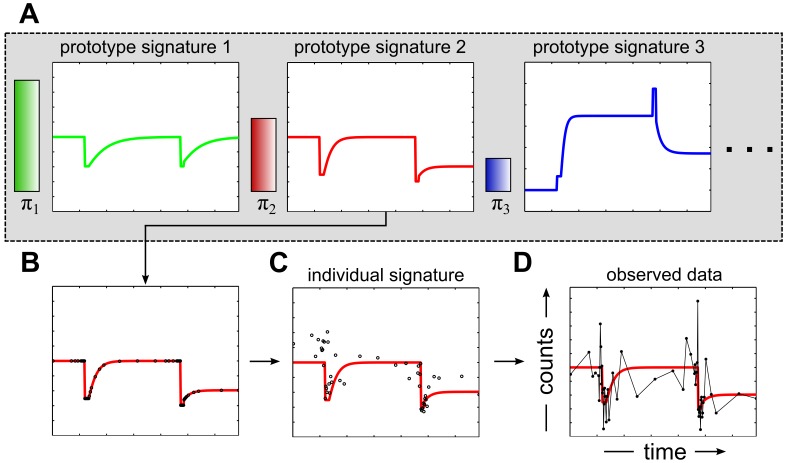
Figure 1. Schematic of the Microbial Counts Trajectories Infinite Mixture Model Engine (MC-TIMME) generative probabilistic model.
Observed data of time-series of sequencing counts for reference operational taxonomic units (refOTUs) are assumed to arise from a multi-level generative probabilistic mixture model.(A)Infinite mixture over latent prototype signatures (green, red and blue solid lines),which specify models of dynamics continuous in both time and amplitude. The horizontal axis for each prototype signature represents time, and the vertical axis represents amplitude. Prototype signatures may adapt their dimensionality, which is shown increasing from left to right. The variables πi and associated shaded bars represent prior probabilities for choosing among prototype signatures. (B)For each refOTU, a prototype signature is probabilistically chosen and sampled at discrete observed time-points. (C)Experiment and refOTU specific variables are added to the selected prototype signature to create an individual signature. (D) Observed data, consisting of sequencing counts, is generated through a discrete-valued noise model parameterized by individual signatures generated in step C.