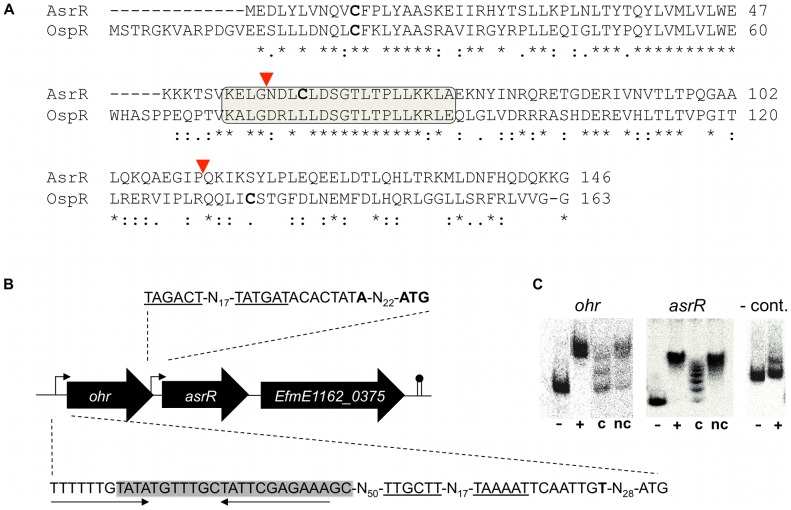
Figure 1. Deduced structure, genomic environment, and DNA binding of AsrR.
(A) Comparison of E. faecium AsrR and P. aeruginosa OspR (Accession number NP_251515.1) amino acid sequences. Asterisks, colons and periods indicate identical, strongly similar, and weakly similar residues, respectively. The winged helix-turn-helix motif is boxed. Cysteine residues, found in AsrR and OspR sequences, are indicated in bold-face characters. The region comprised between red triangles corresponds to the deletion of AsrR protein (ca. 42%) in the ΔasrR strain. (B) Genetic context of the asrR locus. Broken arrows indicate the mapped promoters, start codons and transcription start sites in HM1070 are indicated in bold-face characters, conserved −10 and −35 motifs are underlined. The AsrR binding box, identified experimentally by footprinting (Figure S1C), is boxed on the ohr promoter sequence and putative inverted repeats are highlighted by arrows. The ORF number is indicated according to the strain E1162 annotation (Genbank accession ABQJ00000000). (C) Gel shift experiments showing binding of His6-AsrR to the ohr and asrR promoters. Binding of AsrR was evaluated without (−) or with (+) purified His6-tagged AsrR protein. Specificity of the interaction was evaluated using a control DNA (-cont.) amplified from a non-promoter region and in the presence of unlabelled competitor (c) or non-competitor DNA (nc).