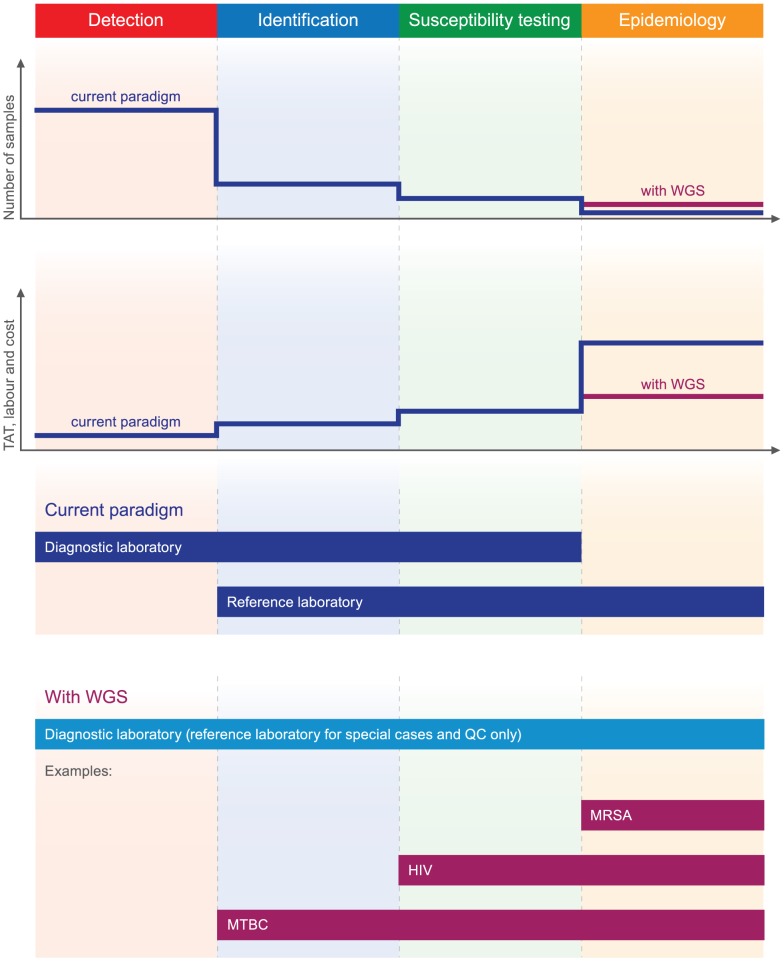
Figure 1. Potential impact of whole genome sequencing in diagnostic and public health microbiology.
Clinical samples processed by diagnostic microbiology laboratories commonly pass through up to four stages, characterised by a stepwise decrease in the number of samples analysed at each successive stage and an inverse association with turn-around time (TAT), labour, and costs (shown in dark blue). Detection, identification, and susceptibility testing in virology are achieved using serological or molecular methods, whereas bacteriology generally relies on phenotypic methods. Epidemiological typing is only done for a handful of organisms using molecular and sometimes phenotypic methods. The most compelling immediate applications for WGS are molecular epidemiology for the purposes of surveillance and outbreak investigation (e.g. for MRSA) and drug susceptibility testing for organisms that are either slow growers or difficult to culture (e.g. MTBC and HIV). This is likely to lead to more samples being typed than is currently the case (depicted in purple), all while drastically reducing turnaround times, provided that WGS is performed in a regional or local laboratory rather than a reference centre. Assuming that this information enables cost-effective clinical interventions, the number of pathogens sequenced is likely to increase over time. Approaches to detection and identification of pathogens and the majority of susceptibility testing are likely to remain largely unchanged in the near future.