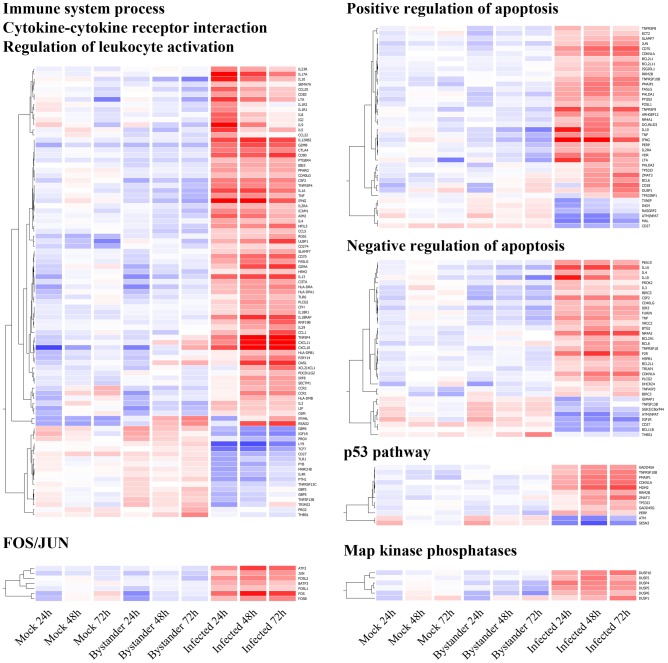
Figure 3. Gene Ontology overrepresentation analysis of genes differentially expressed in HIV-1-infected cells.
Unsupervised hierarchical clustering (per gene) of statistically over-represented Gene Ontology & KEGG pathway categories according to DAVID analysis. Since the first three categories (Immune system process – p = 1.9×10−17, Cytokine-cytokine receptor interaction – p 1×10−29, Regulation of leukocyte activation – p = 1.4×10−9) overlap significantly, they are presented as a single cluster. Positive and negative regulation of apoptosis (p = 5.8×10−7 and 6×10−6, respectively) are both part of the Regulation of apoptosis category, but are here presented separately to better illustrate the absence of trend, in either direction. FOS/JUN (p = 2.3×10−5), the p53 pathway (p = 2.2×10−6) and Map kinase phosphatase (p = 2.3×10−5) are also identified as being over-represented and have implication for HIV-1 biology. All p-values mentioned above are corrected for multiple testing according to Bonferroni. The same color scale as Figure 2 applies.