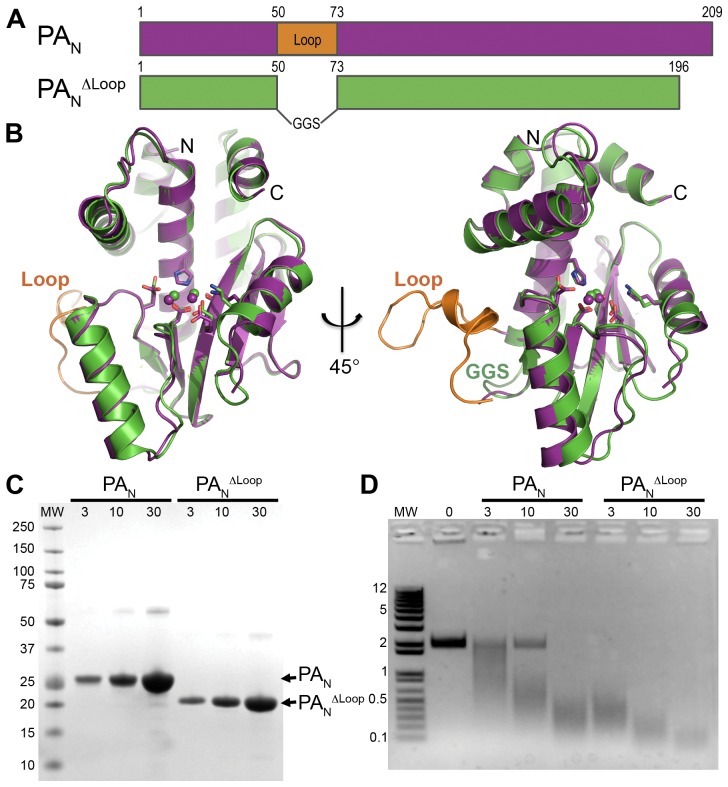
Figure 1. Crystal structure and endonuclease activity of PAN ΔLoop.
(A) Schematic of PAN (magenta) and PAN ΔLoop (green), and location of the 22-residue loop (orange) replaced by a Gly-Gly-Ser linker in the PAN ΔLoop construct. (B) Two orthogonal views of the overlay of the crystal structures of PAN and PAN ΔLoop, colored as in A. Key active site residues are shown in stick representation, the paired manganese ions are shown as spheres, and the N- and C-termini are labeled. The coordinates for PAN are from PDB entry 2W69. The atomic coordinates and structure factors for PAN ΔLoop have been deposited in the Protein Data Bank as PDB entry 4E5E. (C) Coomassie-stained SDS-PAGE of PAN and PAN ΔLoop showing the amount of protein (µM) in a 10 µl endonuclease activity assay reaction. Molecular weight (MW) markers (kD) are shown on the left. (D) Endonuclease activity assay with PAN and PAN ΔLoop. Single-stranded DNA plasmid M13mp18 was incubated with increasing concentrations (µM) of PAN or PAN ΔLoop. Reactions products were resolved on a 1.0% agarose gel stained with ethidium bromide. Molecular weight (MW) ladder (kb) is shown on the left.