Abstract
Members of the fungal genus Fonsecaea causing human chromoblastomycosis show substantial geographic structuring. Genetic identity of clinical and environmental strains suggests transmission from plant debris, while the evolutionary processes that have led to spatially separated populations have remained unexplained. Sequences of ITS, BT2, ACT1, Cdc42, Lac and HmgA were analyzed, either by direct sequencing or by cloning. Thirty-seven clinical and environmental Fonsecaea strains from Central and South America, Asia, Africa and Europe were sequenced and possible recombination events were calculated. Phylogenetic trees of Cdc42, Lac and HmgA were statistically supported, but ITS, BT2 and ACT1 trees were not. The Standardized Index of Association (IA S) did not detect recombination (IA S = 0.4778), neither did the Phi-test for separate genes. In Fonsecaea nubica non-synonymous mutations causing functional changes were observed in Lac gene, even though no selection pressures were detected with the neutrality test (Tajima D test, p>0.05). Genetic differentiation of populations for each gene showed separation of American, African and Asian populations. Strains of clinical vs. environmental origin showed genetic distances that were comparable or lower than found in geographic differentiation. In conclusion, here we demonstrated clonality of sibling species using multilocus data, geographic structuring of populations, and a low functional and structural selective constraint during evolution of the genus Fonsecaea.
Introduction
The genus Fonsecaea comprises etiologic agents of human chromoblastomycosis, a chronic (sub)cutaneous infection eventually leading to cauliflower-like eruptions on the skin [1], [2]. The fungus is present in human tissue in the form of muriform cells. The disease has been reported worldwide, but mostly in tropical and subtropical climate zones, with high incidence in endemic areas [3]–[7].
Inoculation of contaminated thorns or wooden splinters has been hypothesized to be a main route of infection [8], [9]. Thus far the etiologic agents within Fonsecaea are limited to three closely related siblings composing a clearly delimited clade [10]: Fonsecaea pedrosoi, F. monophora and F. nubica. Environmental sampling to recover the species from their supposed natural habitat has been done [8], [9]. F. pedrosoi and F. monophora were only rarely encountered. However, the majority of Fonsecaea-like strains concerned non-virulent species, which were not frequently isolated from on human infections [8]. Either the natural habitat of pathogenic Fonsecaea species has to be found somewhere else, or, alternatively, the species have some kind of advantage of being carried by a mammal host. The existence of evolutionary processes supporting the latter hypothesis may be revealed by comparing patterns of variability and distribution of potential etiologic agents.
The pathogenic strains form a well-supported clade in the Chaetothyriales [11], but specific delimitation within this clade is still a debated issue. Analysis of global genetic diversity using AFLP showed that five groups were distinguishable, which were considered to belong to three different species. Fonsecaea pedrosoi was relatively homogeneous and was found nearly exclusively in Central and South America, while F. monophora and F. nubica each comprised several AFLP groups and had worldwide distribution. Cases were found in a tropic climate zone around the equator, while the few clinical cases outside endemic areas were supposed to have been distributed by recent migration of the human host [11].
In the present study, we investigate patterns of variability of pathogenic Fonsecaea species using multilocus analysis of five functional genes with anonymous sequence and AFLP markers. The set of strains analyzed comprised clinical and environmental strains from three continents.
Materials and Methods
Ethical Standards
The present study has been fully reviewed and approved by Sun Yat-Sen University’s Academic Committee. All subjects provided written informed consent and the procedures have been approved by the Sun Yat-sen University Medical Ethics Committee.
Fungal Strains and Culture Conditions
Seventeen strains of F. pedrosoi, 12 of F. monophora, 8 of F. nubica (Table 1) and one of a neighbouring Cladophialophora species were obtained from the reference collection of the Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre (Utrecht, the Netherlands), in addition to fresh strains recovered from patients, and environmental isolates. Stock cultures were maintained on slants of 2% malt extract agar (MEA) and oatmeal agar (OA) at 24°C.
Table 1. Detailed information of Fonsecaea isolates used in this study.
| Taxonomicname | CBS number | origin | Host/sex | Location | AFLPgenotyping | Multilocusgenotyping | ||
| Cdc42 | Lac | HmgA | ||||||
| F.nubica | CBS 121733 | Chromoblastomycosis | Human/M | China, Guangdong | A | A | A | A |
| CBS 121720 | Chromoblastomycosis | Human/M | China, Guangdong | A | A | A | A | |
| CBS 121734 | Chromoblastomycosis | Human/M | China, Guangdong | A | A | A | A | |
| CBS 269.64 | Chromoblastomycosis | Human/F | South Africa | ND | B | B | B | |
| CBS 444.62 | Chromoblastomycosis | Human/M | Surinam | ND | B | B | B | |
| CBS 557.76 | Unknown | Unknown | Unknown | B | B | B | B | |
| CBS 270.37 | Unknown | Unknown | France (fromS. America) | B | B | B | B | |
| CBS 277.29 | Chromoblastomycosis | Human/M | Brazil | B | B | B | B | |
| F.monophora | CBS 102243 | Chromoblastomycosis | Human/M | Brazil, Parana,Ibituva | C | C | C | C |
| CBS 117236 | Brain | Human/M | United States | C | C | C | C | |
| CBS 102246 | Chromoblastomycosis | Human/M | Brazil, Parana,Campo Largo | C | C | C | C | |
| CBS 269.37 | Chromoblastomycosis | Human | South America | C | C | C | C | |
| CBS 102238 | Soil | Soil | Brazil, Parana,Tibagi River | C | C | C | C | |
| CBS 102229 | Decaying vegetable cover | Plant | Brazil, Parana,Piraquara | C | C | C | C | |
| CBS 397.48 | Chromoblastomycosis | Human/M | South America | C | C | C | C | |
| CBS 102248 | Chromoblastomycosis | Human/M | Brazil, Parana,Piraquara | C | C | C | C | |
| CBS 121727 | Chromoblastomycosis | Human/M | China, Guangdong | D | D | D | C | |
| CBS 121721 | Chromoblastomycosis | Human/M | China, Guangdong | D | D | D | C | |
| CBS 117238 | Brain | Human | United Kingdom | D | D | D | C | |
| CBS 121724 | Chromoblastomycosis | Human/M | China, Guangdong | D | D | D | C | |
| F.pedrosoi | CBS 273.66 | Mouse passage | Soil | Venezuela | ND | E | E | D |
| CBS 271.37 | Chromoblastomycosis | Human/M | South America | E | E | E | D | |
| CBS 671.66 | Mouse passage | Soil | Venezuela | E | E | E | D | |
| CBS 274.66 | Mouse passage | Soil | Venezuela | E | E | E | D | |
| CBS 102247 | Chromoblastomycosis | Human/M | Brazil, Parana | E | E | E | D | |
| CBS 122740 | Chromoblastomycosis | Human/M | Mexico, Mexico City | E | E | E | D | |
| CBS 122736 | Chromoblastomycosis | Human/M | Mexico, Mexico City | E | E | E | D | |
| CBS 122849 | Chromoblastomycosis | Human/M | Mexico, Mexico City | E | E | E | D | |
| CBS 285.47 | Chromoblastomycosis | Human/M | Puerto Rico | E | E | E | D | |
| CBS 342.34 | Chromoblastomycosis | Human/M | Puerto Rico | E | E | E | D | |
| CBS 122741 | Chromoblastomycosis | Human/M | Mexico, Mexico City | E | E | E | D | |
| CBS 670.66 | Mouse passage | Soil | Venezuela | E | E | E | D | |
| CBS 212.77 | Chromoblastomycosis | Human/M | Netherlands,Amsterdam | E | E | E | D | |
| CBS 117910 | Chromoblastomycosis | Human/M | Venezuela, Coro,Falcón State | E | E | E | D | |
| CBS 272.37 | Chromoblastomycosis | Human | Brazil | E | E | E | D | |
| CBS 253.49 | Chromoblastomycosis | Human | Uruguay,Montevideo | E | E | E | D | |
| CBS 201.31 | Gazelle, ear | Animal | Libya, Cyrenaica,Derna | E | E | E | D | |
| Cladophialophora sp. | CBS 109631 | Unknown | Human | Uruguay | F | F | F | E |
CBS: Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre, Utrecht, Netherlands.
ND: not determined.
DNA Extraction and Identification
DNA extraction and quality test were performed as previously reported [12], [13]. DNA concentrations were measured with nano-drop DNA concentration detector at 260 nm (Thermo Scientific, U.S.A.).
Degenerate Primer Design, Cloning and Specific Primer Design for Cdc42, Lac and HmgA
Degenerate and specific primers of Cdc42 refer to the study of Xie et al. [14]. The degenerate primers of HmgA and Lac were designed using a complete alignment of the amino acid sequences of species listed in Table 2. Multiple sequence alignments were generated with the software Clustal W [15] using the amino acid substitution matrix BLOSUM62 [16], [17]. Highly conserved areas were chosen for degenerate primer design. Degenerate forward and reverse primers were designed with minimal degenerate degree using Primer 5.0 software (Table 2).
Table 2. Degenerate primers and specific primers used in this study.
| Degenerate primers | ||||
| Gene | primer | Amino acid sequences | Degenerate nucleotide sequences | |
| Cdc42 | Cdc42-F | G K T C L L I S | GGR AAR ACM TGY YTN ATH TCN TC | |
| Cdc42-R | L K D V F D E A | GCC TCR TCR AAR ACW KYC TTS A | ||
| Lac | Lac-Ds | V T H C P I P | GTK ACD CAR TGY CCS ATT CC | |
| Lac-Das | H G H V H P P | TG SCC RTG VAR RTG GAA CGG | ||
| HmgA | HmgA-F2 | F T A P R H E | TTY ACN GCN CCN MGN CAY GA | |
| HmgA-R12 | N H G N Y Y P | GG RTA RTA RTT NCC RTG CC | ||
| HmgA-R22 | P P N Y H R N | TT NCK RTG RTA CCA NGG NGG | ||
| Specific primers | ||||
| Gene | primer | Specific nucleotide sequences | Reference | |
| Cdc42 | Cdc42-SF1s | GGC AAG ACA TGC TTG TTG ATC TC | This study | |
| Cdc42-SR1s | GCC TCG TCA AAT ACG TCC TTA A | |||
| Lac | LacIs | CGC CAG GCT TTG ATT GTG | This study | |
| LacIas | CGC CGT CGT TAT TGT TGA G | |||
| HmgA | HmgA-F2s | TTR ACT GCG CCA CGR CAC GA | This study | |
| HmgA-R12s | GG RTA RTA RTT GCC RTG CCA T | |||
| ITS | V9G, LS266 | Masclaux et al. (1995) | ||
| BT2 | Bt2a, Bt2b | White et al. (1990 | ||
| ACT1 | Actaw, Actfw | Glass & Donaldson (1995) | ||
DNA of type strains of the genus Fonsecaea were used as the PCR amplification template. Optimal amplification condition was optimized by temperature gradient PCR amplification. Specific amplicons were purified using gel extraction kit (Qiagen, Germany), cloned using a cloning kit (Promega, Madison, WI, U.S.A.) and confirmed by direct PCR amplification with the primer set M13fw (5′-GTA AAA CGA CGG CCA GT-3′) and M13rv (5′-GGA AAC AGC TAT GAC CAT G-3′) according to the manufacturer’s instructions. PCR amplicons were then purified with Sephadex G-50 fine (GE Healthcare Bio-Sciences, Uppsala, Sweden) and sequencing was done on an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA, U.S.A.). Sequence data were edited using the SeqMan of Lasergene software (DNAStar, Madison, WI, U.S.A.). The resulting sequences were aligned using BioNumerics software v. 4.61 (Applied Maths, Kortrijk, Belgium). The specificity of these sequences for three genes was confirmed by BLASTx search on GenBank (http://blast.ncbi.nlm.nih.gov). Specific primers for HmgA and Lac were obtained by comparison with the degenerate primer of HmgA and Lac respectively. The resulting specific primers cdc42-SF1s, cdc42-SR1s, Lac-Is, Lac-IAs, HmgA-F2s and HmgA-R12s (Table 2) were subsequently tested with the aim to establish amplification conditions, and then were used to test the 38 strains listed in Table 1.
Multilocus Gene Amplification and Sequencing
PCR amplification and sequencing of ITS, BT2, ACT1 was done according our earlier study [18]. PCR amplification of Cdc42, Lac and HmgA was performed with cdc42-SF1s and cdc42-SR1s, LacIS and LacIAS, and HmgA-F2s, HmgA-R12s and HmgA-R22s, respectively. PCR was performed in a 50 µl volume of a reaction mixture containing 14 µl Go Taq master mix (Promega) containing dNTPs, MgCl2, reaction buffer, 2 µl of each primer (10 pmol) and 1 µl DNA. Amplification was performed in an ABI PRISM 2720 (Applied Biosystems) thermocycler as follows: 95°C for 5 min, followed by 35 cycles consisting of 95°C for 45 sec, 49.5°C for 30 sec and 72°C for 1.5 min, and a delay at 72°C for 7 min. Annealing temperature was changed to 52°C for Lac. A seminested PCR was performed to amplify the HmgA gene, the first run with primer HmgA-F2s and HmgA-R22, as follows: 95°C for 5 min, followed by 35 cycles consisting of 95°C for 45 sec, 49.5°C for 30 sec and 72°C for 1.5 min, and a delay at 72°C for 7 min. One µl amplicon of the first run were used as templates for the second run with primer HmgA-F2s and HmgA-R12s under the same reaction conditions. Sequencing of PCR amplicons was done on an ABI 3730XL automatic sequencer (Applied Biosystems, U.S.A.). Sequence data were edited using the SeqMan of Lasergene software (DNAStar Inc., Madison, U.S.A.).
Phylogenetic Reconstruction and DNA Polymorphism
The Cipres Portal (http://www.phylo.org) was used to construct maximum likelihood trees with RAxML v. 7.2.6 for ITS, BT2, ACT1, Cdc42, Lac and HmgA. Maximum likelihood searches for the best scoring tree were made after a bootstrap estimate of the proportion of invariable sites automatically determined the number of bootstrapping runs. RAxML will then automatically determine the point at which enough bootstrapping replicates have been produced [19]. Bootstrap values equal to or greater than 80% were considered significant. After repeated construction for all six markers, the combined single file was used to calculate the standardized Index of Association, IA S [20] using the Lian 3.5 webserver (http://pubmlst.org). The test options were set to Monte Carlo with 1,000 iterations/random resamplings. The same alignments were used to show split-decomposition trees using Splitstree 4 v. 4.8. The same software package was used to apply Phi test (pairwise homoplasy index) to distinguish recurrent mutations (or homoplasies) from recombination in generating genotypic diversity.
DNA polymorphism analyses were carried out using DNASP 5.10.00 software. A subset of Fonsecaea strains and genotypes was used to calculate haplotype and nucleotide diversity, as well as Tajima’s D neutrality test that is based on the number of pairwise differences and the number of segregating sites in a sample of sequences and the number of parsimonious informative sites [21]. The same software package was used to calculate FST, showing the genetic differentiation among populations, for ITS, BT2, ACT1, Cdc42, Lac and HmgA.
AFLP Genotyping Assay
AFLP genotyping data were taken from our previous study, where a detailed description of the methodology is provided [11].
Laccase and Homogentisate 1,2-dioxygenase Enzyme Activity Assays
All strains representing F. pedrosoi, F. monophora and F.nubica indicated in Table 1 were tested for laccase and homogentisate 1, 2-dioxygenase enzyme activities. Tests were repeated three times for each strain. Laccase was tested according to Mander et al. [22]. Solid MM with a pH of 5 supplemented with 5 mM 2, 2-azino-di-(3-ethylbenzthiazolinsulfonate) (ABTS) which is oxidized by laccase and results in colored compounds. Cultures were pre-incubated at 25°C for 7 days. Subcultures were cut with a cork borer 2 mm diam and placed at the centre of the plate with three replicates. Diameters of colored metabolite halos were measured from day 1 to day 7. For the homogentisate 1,2-dioxygenase enzyme activity test, we followed Ye & Szaniszlo [23]. Solid MM was supplemented separately with 5 mM L-phenylalanine (Sigma, U.S.A.) and 5 mM L-tyrosine (Sigma) which served as artificial substrates to evaluate the homogentisate 1,2-dioxygenase enzyme activity. Culture conditions were the same as in the laccase test. After two weeks of culture, colony diameters were measured.
Statistics
Metabolite diameters were analyzed by one way ANOVA using Prism 5.0 software, followed by Tukey’s HSD Post-hoc test. Mean diameters are the result of triplicate experiments. The error bars indicate standard error of the mean; p<0.05 was considered to indicate a significant difference.
Results
Primer Development for Cdc42, Lac and HmgA
Highly conserved domains were found in Cdc42, HmgA and Lac genes after comparison of sequences downloaded from GenBank (Table 3) and these were used for degenerate primer design. PCRs with degenerate primer pairs Cdc42-F and Cdc42- R, Lac-Ds and Lac-Das, HmgA-F2 and HmgA-R12/HmgA-R22 yielded multiple bands. After cloning and alignment analysis, the specific primers cdc42-SF1s and cdc42-SR1s, Lac-IS and Lac-IAS, and HmgA-F2s, HmgA-R12s/HmgA-R22s were obtained (Table 2). The sets of specific primers each yielded single PCR products of about 0.85 kb, 1 kb and 0.9 kb, respectively (data not shown). The introns were taken out when used for further analysis. The primer sets proved to amplify all Fonsecaea agents of chromoblastomycosis successfully. To establish an outgroup, degenerate primers were used to amplify the target gene, and multiple bands were cloned and sequenced. Blast searches using translated amino acid sequences in GenBank showed that the amplified fragments of Cdc42, Lac and HmgA had high homology with published target genes [24]–[26]. The conserved domain search revealed that Cdc42 contains a Ras-like GTPase superfamily (aa1–120) which involved a GTP/Mg2+ binding site (aa45–100) and switch I and II regions (aa20–25, aa40–60) [27]. Lac contained a Cu-oxidase superfamily which typically exists in the laccase family [27]. HmgA contained the HgmA superfamily (aa1–204), a hexamer arrangement consisting of a dimer of trimers with which the active site iron ion is coordinated [27].
Table 3. Homogentisate 1,2-dioxygenase (HmgA) and laccase (Lac) references taken from GenBank.
| Taxonomic name | Associated strain number | gene | GenBank no. protein |
| Ajellomyces dermatitidis | SLH14081 | HmgA | XP_002626277.1 |
| Trichophyton tonsurans | CBS 112818 | HmgA | EGD98945 |
| Coccidioides immitis | RS | HmgA | XP_001247541.1 |
| Paracoccidioides brasiliensis | Pb03 | HmgA | EEH17396.1 |
| Trichophyton tonsurans | CBS 112818 | HmgA | EGD98945.1 |
| Trichophyton equinum | CBS 127.97 | HmgA | EGE07801.1 |
| Aspergillus terreus | NIH2624 | HmgA | XP_001218689.1 |
| Aspergillus niger | CBS 513.88 | HmgA | XP_001388730.2 |
| Aspergillus oryzae | RIB40 | HmgA | XP_001727215.2 |
| Trichophyton rubrum | CBS 118892 | HmgA | XP_003238076.1 |
| Neurospora crassa | OR74A | HmgA | XP_960461.1 |
| Aspergillus fumigatus | Af293 | HmgA | XP_750969.1 |
| Penicillium marneffei | ATCC 18224 | HmgA | XP_002150285.1 |
| Neurospora crassa | Lac | AAA33591.1 | |
| Cryptococcus neoformans var. grubii | Lac | ABI58272.1 | |
| Cryptococcus neoformans var. neoformans | JEC21 | Lac | AAW46742.1 |
| Aspergillus nidulans | FGSC A4 | Lac | XP_664239.1 |
| Aspergillus flavus | NRRL3357 | Lac | EED57644.1 |
| Aspergillus terreus | NIH2624 | Lac | EAU34323.1 |
| Talaromyces stipitatus | ATCC 10500 | Lac | EED19078.1 |
| Ajellomyces dermatitidis | SLH14081 | Lac | XP_002629368.1 |
| Penicillium marneffei | ATCC 18224 | Lac | EEA21273.1 |
| Aspergillus clavatus | NRRL 1 | Lac | EAW07265.1 |
| Aspergillus fumigatus | Af293 | Lac | XP_752933.1 |
| Trichophyton tonsurans | CBS 112818 | Lac | EGD95875.1 |
| Coccidioides immitis | RS | Lac | XP_001239516.1 |
CBS: Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre, Utrecht, Netherlands.
NIH: The National Institute of Heath, Bethesda, Maryland, USA.
ATCC: American Type Culture Collection, Manassas, VA, USA.
FGSC: The Fungal Genetics Stock Center, Kansas City, Missouri, USA.
NRRL: ARS Culture Collection, Washington DC, USA.
Phylogeny
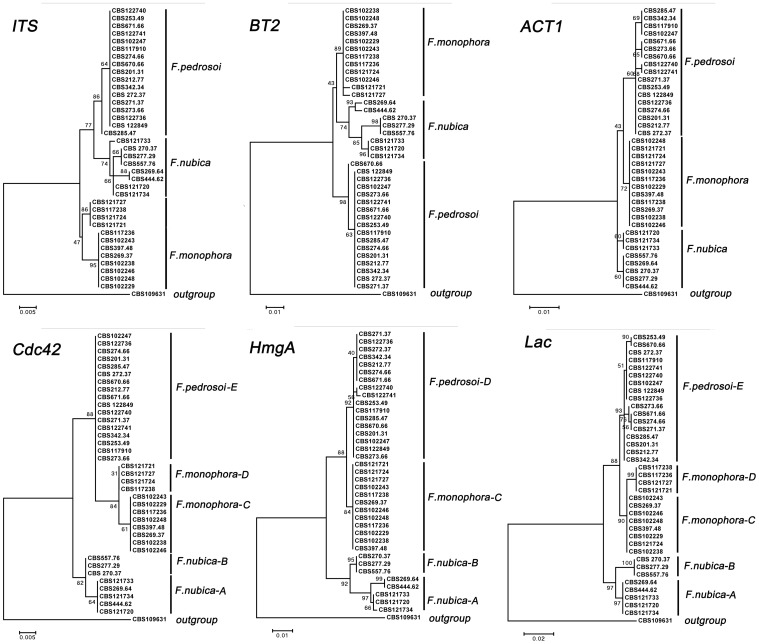
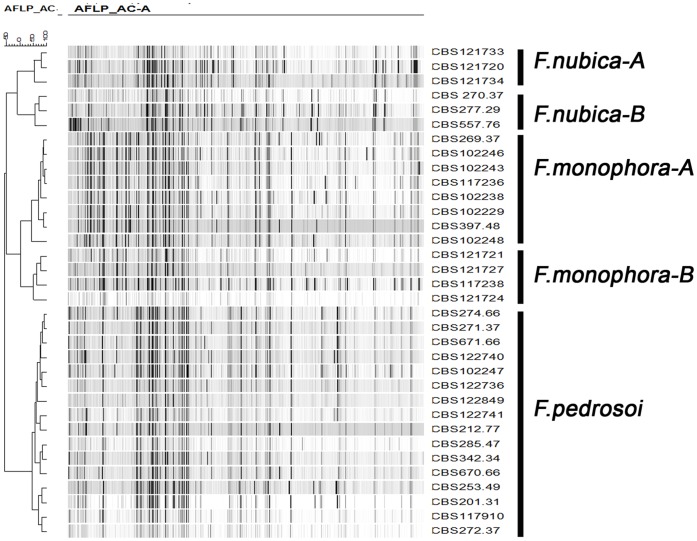
Six phylogenetic trees were constructed for 37 Fonsecaea strains distributed globally using sequenced ITS, BT2, ACT1, Cdc42, Lac and HmgA genes, and one Cladophialophora strain (CBS 109631) used as outgroup. Three clades corresponding to F. pedrosoi, F. monophora and F. nubica showed strong support in Cdc42, Lac and HmgA genes (bootstrap values >80%) (Fig. 1). F.pedrosoi showed limited variability within the species. Two subclades were distinguished within F. monophora with high bootstrap support in Cdc42 and Lac (Fig. 1), while within F.nubica, two subclades in Cdc42, Lac and HmgA (Fig. 1). The AFLP genotyping assay showed the similar tree topology (Fig. 2), with five subclades with high bootstrap support within the genus Fonsecaea. However, for ITS, BT2 and ACT1 genes, no significant bootstrap support was obtained (Fig. 1). Fixed populations were observed throughout the six phylogenetic and AFLP genotyping trees. The F. pedrosoi clade comprised 17 strains from patients and from the environment in South America and Europe. F.monophora genotype A comprised 4 clinical strains from South China, and genotype B comprised 8 strains from patients and the environment in South America. F.nubica genotype A comprised 3 clinical strains from in Europe and South America, and genotype B comprised 5 clinical strains from Africa and China (Table 1, Fig. 1).
Figure 1. Consensus trees of Fonsecaea based on ITS ribosomal DNA, BT2, ACT1, Cdc42, HmgA and Lac of 37 strains, constructed with MEGA5.0 and 500 bootstrap replicates, CBS 109631 was taken as outgroup.
Figure 2. Clustering of amplified fragment-length polymorphism banding pattern of isolates of Fonsecaea spp. analyzed by using unweighted pair group method with arithmetic mean.
Subclusters showed as in Figure.
Multilocus Recombination Analyses
Standardized Index of Association IA S [28] performed using Lian 3.5 [20] confirmed clades according to the RaxML trees in the different partitions. IA S measures the degree of association between alleles at different loci based on the variance in genetic distance between genotypes and is expected to be 0 if populations are freely recombining and >0 if there is an association between alleles. The calculated index using 1000 Monte Carlo resamplings was 0.4778 (VD = 3.8561, Ve = 0.9973), showing no evidence of recombination. The Phi-test [29] is performed for individual loci for detection of recombination within sequences with expected recombination events the value is p<0.05, otherwise, homoplasy (p>0.05) is considered. The results based on six genes showed no significant statistical evidence of recombination at ITS (p = 0.076), BT2 (p = 0.112), ACT1 (p = 1.0), Cdc42 (p = 1.0), Lac (p = 0.79), or HmgA (p = 0.46).
Strain Polymorphism Statistics
The calculated parsimonious informative sites, monomorphic sites, segregating sites and the total number of mutations are summarized in Table 4. In total 37 strains were used for all six genes. The haplotype diversity for ITS (0.761), BT2 (0.743), ACT1 (0.824), Cdc42 (0.725), Lac (0.883) and HmgA (0.820) were in comparable range. For the neutrality test, Tajima’s D values for ITS (1.21111), BT2 (0.27942), ACT1 (0.65057), Cdc42 (1.23506), Lac (0.63696) and HmgA (0.61812) were not statistically supported (p>0.10), no positive selection being detected within the tested genus (Table 4).
Table 4. Phylogenetic marker diversity and molecular evolutionary parameters for the gene segments examined.
| Parameters | Phylogenetic Marker | ||||||
| ITS | Cdc42 | Lac | HmgA | BT2 | ACT1 | ||
| Fragment features | Exon/intron | Exon | Exon | Exon | Exon/intron | Exon/intron | |
| No. of sequences | 37 | 37 | 37 | 37 | 37 | 37 | |
| No. of characters | 572 | 360 | 708 | 612 | 303 | 486 | |
| No. of codon | n.a | 120 | 236 | 204 | 83 | 144 | |
| DNA polymorphism analysis | |||||||
| Gaps/missing data | 572 | 360 | 708 | 612 | 278 | 485 | |
| Segregating sites | 22 | 8 | 37 | 32 | 18 | 10 | |
| No. of mutations (η) | 22 | 8 | 38 | 32 | 18 | 10 | |
| No. of haplotypes | 6 | 5 | 9 | 7 | 9 | 7 | |
| Haplotype diversity | 0.761 | 0.725 | 0.883 | 0.820 | 0.743 | 0.824 | |
| Nucleotide diversity | 0.01417 | 0.00662 | 0.01478 | 0.01374 | 0.01682 | 0.00600 | |
| Neutrality analysis | |||||||
| Tajima’s D test | 1.21111 (p>0.10) | 1.23506 (p>0.10) | 0.63696 (p>0.10) | 0.61812 (p>0.10) | 0.27942 (p>0.10) | 0.65057(p>0.10) | |
The values of FST lie between 0 (panmictic) and 1 (total separation). The tested FST values based on six genes by comparing geographic origins of the strains (Table 5A) show similar values between South and Central America, while those of Chinese and Africa strains were higher. The FST values based on six combined genes from clinical (28 strains) and environmental origins (6 strains) (Table 1) showed a comparable or lower value (0.07567) than with comparisons of geographic origins between South America and Central America (0.10549), South America and Africa (0.33106), South America and Asia (0.25447), Central America and Africa (0.41542), Central America and Asia (0.425565), and Asia and Africa (0.17738) (Table 5B). The comparisons of geographic origins between continents low values were found (Table 5) suggesting separation of populations.
Table 5. Population differentiation index (FST) of 37 Fonsecaea strains based on separate (A) and combined (B) multilocus gene sequences clustered by geographical origin. Seven strains from China, 3 from Africa, 17 from South America and 6 from Central America.
| A | South America | Central America | Africa | |||||||||||||||
| ITS | ACT1 | BT2 | Cdc42 | HmgA | Lac | ITS | ACT1 | BT2 | Cdc42 | HmgA | Lac | ITS | ACT1 | BT2 | Cdc42 | HmgA | Lac | |
| CA | 0.0865 | 0.1304 | 0.1727 | 0.0768 | 0.0875 | 0.0788 | ||||||||||||
| AF | 0.2658 | 0.2148 | 0.2524 | 0.3874 | 0.2547 | 0.3158 | 0.4755 | 0.3323 | 0.4352 | 0.4640 | 0.3360 | 0.4802 | ||||||
| AS | 0.1890 | 0.2161 | 0.2286 | 0.1638 | 0.1437 | 0.1664 | 0.3989 | 0.5230 | 0.5764 | 0.3121 | 0.3894 | 0.3533 | 0.2341 | 0.2237 | 0.2378 | 0.1536 | 0.0187 | 0.0821 |
| B | South America | Central America | Africa | |||||||||||||||
| CA | 0.10549 | |||||||||||||||||
| AF | 0.33106 | 0.41542 | ||||||||||||||||
| AS | 0.25447 | 0.425565 | 0.17738 | |||||||||||||||
Synonymous and non-synonymous changes of the genus Fonsecaea in amino acid sequence in six genes are listed in Table 4. In total 787 amino acid codons were used for the comparison, and 8 1st base, 2 2nd base and 81 3rd base mutations were found within the three species. All 1st base mutations caused non-synonymous changes, but the 2nd base and 3rd base mutations caused synonymous changes. A further analysis showed that the non-synonymous changes in ACT1 and BT2 both did not occur in the functional domain (ACT1aa135, BT2aa81), while non-synonymous changes in Lac and HmgA both occurred in functional domains (Lacaa159, HmgAaa38, aa88, aa164, aa175). Most non-synonymous changes were observed in F. nubica, where all strains isolated to date originate from chromoblastomycosis patients (Table 6).
Table 6. Synonymous and non-synonymous changes in DNA and amino acid sequence in ACT1, BT2, Cdc42, Lac and HmgA genes of Fonsecaea spp.
| Gene | Species | Total codon | 1st base | 2ed base | 3rd base | Amino acid change | Strains |
| ACT1 | F. pedrosoi | 144 | |||||
| F. monophora | 144 | ||||||
| F. nubica | 144 | 1 | 9 | CAT→TAT/H→Y | All tested F. nubica | ||
| BT2 | F. pedrosoi | 83 | 1 | 3 | TAT→GAT/Y→D | CBS 671.66, CBS 273.66, CBS 670.66 | |
| F. monophora | 83 | 1 | |||||
| F. nubica | 83 | 1 | |||||
| Cdc42 | F. pedrosoi | 120 | |||||
| F. monophora | 120 | 4 | |||||
| F. nubica | 120 | 4 | |||||
| Lac | F. pedrosoi | 236 | 1 | 5 | |||
| F. monophora | 236 | 7 | |||||
| F. nubica | 236 | 1 | 1 | 22 | CCG→CTG/P → L | All tested F. nubica | |
| HmgA | F. pedrosoi | 204 | 1 | ||||
| F. monophora | 204 | 3 | |||||
| F. nubica | 204 | 4 | 1 | 21 | AGC→GGC/S→GGCC→ACC/A→TAGC→AAC/S→NGCT→ACT/A→T | All tested F. nubicaAll tested F. nubicaAll tested F. nubicaAll tested F. nubica | |
| Total | 787 | 8 | 2 | 81 |
H: Histidine, Y: Tyrosine, D: Ariginine, P: Proline, L: Leucine, S: Serine, G: Glycine, A: Alanine, T: Threonine, N: Asparagine.
CBS: Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands.
Laccase and Homogentisate 1,2-dioxygenase Enzyme Activity Assay
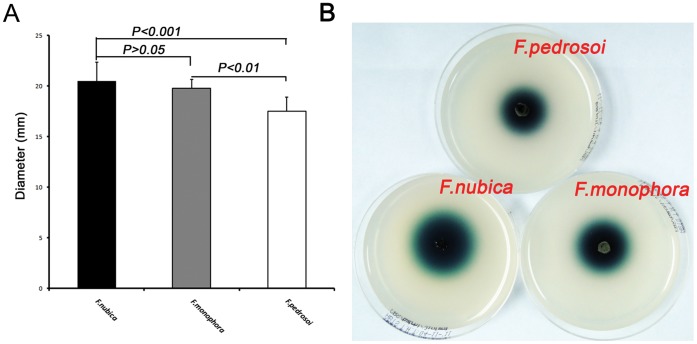
All strains tested yielded positive laccase activity. Colored metabolites were observed in all three species, but statistical analysis showed that F. nubica had higher enzyme activity than other species (F. nubica vs. F. pedrosoi, p<0.001, F. nubica vs. F. monophora, p>0.05, F. monophora vs. F. pedrosoi, p<0.01) (Fig. 3). The homogentisate 1,2-dioxygenase enzyme activity assay revealed that all strains are able to assimilate L-phenylalanine and L-tyrosine as sole carbon sources; no difference was observed within the three species (data not shown).
Figure 3. Laccase activity assay.
Colored metabolite diameters of tested strains were measured after 7-day culture at 25°C on solid MM medium with 5 mM ABTS. Statistical analysis shown as mean ± standard deviation (A). The plates show the generation of colored metabolite compound (B), F. pedrosoi (CBS 273.66), F. monophora (CBS 117236), F. nubica (CBS 121720).
Discussion
In the evolution of black fungi (order Chaetothyriales) [30], we witness a functional change from a rock-inhabiting life style prevalent in ancestral Coniosporium (Knufia) and relatives to an increased ability to infect humans and other vertebrates in derived clades. Agents of chromoblastomycosis are particularly interesting because they exhibit a pathogenic phase in tissue, the muriform cell, which shows morphogenetic resemblance isodiametrically enlarging cell clumps of rock-inhabiting Coniosporium (Knufia) species. A functional change in the Cdc42 gene, involved in cellular polarity has been hypothesized [31]. The change of life style seems to have been quite successful in the F. pedrosoi clade, judging from the fact that three related species are nearly exclusively found on humans [32]. Nevertheless the shift was not seen to be reflected in the cytoskeleton-associated Cdc42 gene when compared over the order Chaetothyriales [33].
In the present study, six genes were compared in human-pathogenic Fonsecaea species. ITS was used as a standard for phylogenetic construction. ACT1, BT2 and Cdc42 play a role in cell cycle progression and actin cytoskeleton construction, and are involved in morphogenetic switching, leading to large spherical cells with subsequent cellular division giving rise to the infective muriform cell [34]. Lac and HmgA are well-documented virulence factors of black fungi, and participate in the synthesis of melanin. DHN melanin is negatively charged, hydrophobic and of high molecular weight, and arises by the oxidative polymerization of phenolic and/or indolic precursors [35]. Melanin enhances virulence in black fungi of the order Chaetothyriales [36]–[42]. We developed primers to amplify Cdc42, Lac and HmgA which proved to be specific for Fonsecaea. The sequenced genes were aligned and confirmed to be Cdc42, Lac and HmgA using BLAST oine search in GenBank. The genes contained the gene-specific conserved domains when searched with translated amino acid sequences [27].
The phylogenetic trees reconstructed with Cdc42, Lac and HmgA (Fig. 1) yielded high bootstrap support for the three sibling Fonsecaea species, while the ITS, ACT1 and BT2 trees were not supported. The lack of support was probably caused by incomplete lineage sorting, several mutations not having reached fixation. Based on the Standardized Index of Association (IA S) and Phi-test using six genes, no recombination events were detected among the three sibling species. This phenomenon is frequently observed in opportunistic members of Chaetothyriales, where clonality seems to be prevalent [43]. The neutrality test with Tajima’s D yielded no significant results, suggesting that no positive selection was detected in the sequenced genes indicating a low functional and structural selective constraint during evolution.
Relatively low haplotype diversity was observed within the six genes analyzed. A total of 91 fixed synonymous and non-synonymous changes were observed in coding regions. The non-synonymous changes in the cytoskeleton genes ACT1 and BT2 are not responsible for morphogenetic changes [7], [44] among the three species because the mutations occurred outside functional domains. The non-synonymous changes in Lac and HmgA both occurred in functional domains (Lacaa159, HmgAaa38, aa88, aa164, and aa175) (Table 6), but did not cause obvious functional changes when catalysis of substrates was tested in vitro. A possible explanation might be that the non-synonymous mutations did not cause any changes in the three-dimensional structure of the molecule. A systematic alignment of 223 plant and fungi laccase sequences showed that there are four signature sequence regions (L1-4) and 12 housekeeping amino acids [45], while the detected non-synonymous mutations (Lacaa159) in this study occurred between L2 and L3 and do not belong to a conserved region. DNA sequence alignment of HmgA showed that HmgAaa38, aa88, aa164, and aa175 are not located in conserved regions either. Therefore we conclude that the non-synonymous changes within two genes are not linked to functional or structural selective constraints within the genus Fonsecaea. Subsequent studies may reveal whether such changes have occurred in the analyzed genes in ancestral clades, where dramatic changes in life style are supposed to have taken place.
Several studies reported on the molecular epidemiology of the sibling species Fonsecaea [45]. Ribosomal and mitochondrial DNA typing has been used to map the geographic origins of strains [46], [47]. The molecular epidemiology of this genus showed substantial geographic structuring in all species with differences between American, African and Asian populations similar to what has been found by Kawasaki et al. [46] in mtDNA profiles. In conclusion, we demonstrated clonality of sibling species using multilocus data, geographic structuring of populations, and a detected low functional and structural selective constraint during evolution of the genus Fonsecaea.
Funding Statement
This work was supported by National Natural Science Foundation of China (No.U0932009) and The Federation of European Biochemical Societies (FEBS) Chinese European Visiting Fellowships (No.CEVF-00007), and was partly supported by the International Program of Project 985 at Sun Yat-sen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Queiroz-Telles F, Esterre P, Perez-Blanco M, Vitale RG, Salgado CG, et al. (2009) Chromoblastomycosis: an overview of clinical manifestations, diagnosis and treatment. Med Mycol 47: 3–15. [DOI] [PubMed] [Google Scholar]
- 2. Lopez Martinez R, Mendez Tovar LJ (2007) Chromoblastomycosis. Clin Dermatol 25: 188–194. [DOI] [PubMed] [Google Scholar]
- 3. Esterre P, Andriantsimahavandy A, Ramarcel ER, Pecarrere JL (1996) Forty years of chromoblastomycosis in Madagascar: a review. Am J Trop Med Hyg 55: 45–47. [DOI] [PubMed] [Google Scholar]
- 4. Kombila M, Gomez de Diaz M, Richard-Lenoble D, Renders A, Walter P, et al. (1995) [Chromoblastomycosis in Gabon. Study of 64 cases]. Sante 5: 235–244. [PubMed] [Google Scholar]
- 5. Silva JP, de Souza W, Rozental S (1998) Chromoblastomycosis: a retrospective study of 325 cases on Amazonic Region (Brazil). Mycopathologia 143: 171–175. [DOI] [PubMed] [Google Scholar]
- 6. Attapattu MC (1997) Chromoblastomycosis–a clinical and mycological study of 71 cases from Sri Lanka. Mycopathologia 137: 145–151. [DOI] [PubMed] [Google Scholar]
- 7. Xi L, Sun J, Lu C, Liu H, Xie Z, et al. (2009) Molecular diversity of Fonsecaea (Chaetothyriales) causing chromoblastomycosis in southern China. Med Mycol 47: 27–33. [DOI] [PubMed] [Google Scholar]
- 8. Vicente VA, Attili-Angelis D, Pie MR, Queiroz-Telles F, Cruz LM, et al. (2008) Environmental isolation of black yeast-like fungi involved in human infection. Stud Mycol 61: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salgado CG, da Silva JP, Diniz JA, da Silva MB, da Costa PF, et al. (2004) Isolation of Fonsecaea pedrosoi from thorns of Mimosa pudica, a probable natural source of chromoblastomycosis. Rev Inst Med Trop Sao Paulo 46: 33–36. [DOI] [PubMed] [Google Scholar]
- 10. Najafzadeh MJ, Sun J, Vicente V, Xi L, van den Ende AH, et al. (2010) Fonsecaea nubica sp. nov, a new agent of human chromoblastomycosis revealed using molecular data. Med Mycol 48: 800–806. [DOI] [PubMed] [Google Scholar]
- 11. Najafzadeh MJ, Sun J, Vicente VA, Klaassen CH, Bonifaz A, et al. (2011) Molecular epidemiology of Fonsecaea species. Emerg Infect Dis 17: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun J, Zhang J, Najafzadeh MJ, Badali H, Li X, et al. (2011) Melanization of a meristematic mutant of Fonsecaea monophora increases tolerance to stress factors while no effects on antifungal susceptibility. Mycopathologia 172: 373–380. [DOI] [PubMed] [Google Scholar]
- 13. Sun J, Najafzadeh MJ, Vicente V, Xi L, de Hoog GS (2010) Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J Microbiol Methods 80: 19–24. [DOI] [PubMed] [Google Scholar]
- 14. Xie Z, Feng P, Zhang J, Li X, Sun J, et al. (2012) Molecular cloning, characterization and differential expression of Cdc42 in Fonsecaea monophora . Mol Biol Rep 39: 839–844. [DOI] [PubMed] [Google Scholar]
- 15. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henikoff JG, Henikoff S (1996) Using substitution probabilities to improve position-specific scoring matrices. Comput Appl Biosci 12: 135–143. [DOI] [PubMed] [Google Scholar]
- 17. Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Najafzadeh MJ, Gueidan C, Badali H, Van Den Ende AH, Xi L, et al. (2009) Genetic diversity and species delimitation in the opportunistic genus Fonsecaea . Med Mycol 47: 17–25. [DOI] [PubMed] [Google Scholar]
- 19. Polerecky L, Bissett A, Al-Najjar M, Faerber P, Osmers H, et al. (2009) Modular spectral imaging system for discrimination of pigments in cells and microbial communities. Appl Environ Microbiol 75: 758–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haubold B, Hudson RR (2000) LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- 21. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mander GJ, Wang H, Bodie E, Wagner J, Vienken K, et al. (2006) Use of laccase as a novel, versatile reporter system in filamentous fungi. Appl Environ Microbiol 72: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arias-Barrau E, Olivera ER, Luengo JM, Fernandez C, Galan B, et al. (2004) The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol 186: 5062–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye X, Szaniszlo PJ (2000) Expression of a constitutively active Cdc42 homologue promotes development of sclerotic bodies but represses hyphal growth in the zoopathogenic fungus Wangiella (Exophiala) dermatitidis . J Bacteriol 182: 4941–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lyons JI, Newell SY, Buchan A, Moran MA (2003) Diversity of ascomycete laccase gene sequences in a southeastern US salt marsh. Microb Ecol 45: 270–281. [DOI] [PubMed] [Google Scholar]
- 26. Dong F, Zhong Y, Arulanandam B, Zhong G (2005) Production of a proteolytically active protein, chlamydial protease/proteasome-like activity factor, by five different Chlamydia species. Infect Immun 73: 1868–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith JM, Smith NH, O’Rourke M, Spratt BG (1993) How clonal are bacteria? Proc Natl Acad Sci U S A 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sterflinger K (1998) Temperature and NaCl-tolerance of rock-inhabiting meristematic fungi. Antonie Van Leeuwenhoek 74: 271–281. [DOI] [PubMed] [Google Scholar]
- 31. Cooper CR Jr, Szaniszlo PJ (1993) Evidence for two cell division cycle (CDC) genes that govern yeast bud emergence in the pathogenic fungus Wangiella dermatitidis . Infect Immun 61: 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, van den Ende AH, et al. (2008) Biodiversity of the genus Cladophialophora . Stud Mycol 61: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng S, van den Ende AH, Ram AF, Arentshorst M, Graser Y, et al. (2008) Evolution of CDC42, a putative virulence factor triggering meristematic growth in black yeasts. Stud Mycol 61: 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szaniszlo PJ, Karuppayil SM, Mendoza L, Rennard RJ (1993) Cell cycle regulation of polymorphism in Wangiella dermatitidis . Arch Med Res 24: 251–261. [PubMed] [Google Scholar]
- 35. Nosanchuk JD, Casadevall A (2003) The contribution of melanin to microbial pathogenesis. Cell Microbiol 5: 203–223. [DOI] [PubMed] [Google Scholar]
- 36. Morris-Jones R, Gomez BL, Diez S, Uran M, Morris-Jones SD, et al. (2005) Synthesis of melanin pigment by Candida albicans in vitro and during infection. Infect Immun 73: 6147–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaskes S, Tyndall RL (1978) Pigment production by Cryptococcus neoformans and other Cryptococcus species from aminophenols and diaminobenzenes. J Clin Microbiol 7: 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gomez BL, Nosanchuk JD, Diez S, Youngchim S, Aisen P, et al. (2001) Detection of melanin-like pigments in the dimorphic fungal pathogen Paracoccidioides brasiliensis in vitro and during infection. Infect Immun 69: 5760–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Romero-Martinez R, Wheeler M, Guerrero-Plata A, Rico G, Torres-Guerrero H (2000) Biosynthesis and functions of melanin in Sporothrix schenckii . Infect Immun 68: 3696–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nosanchuk JD, Gomez BL, Youngchim S, Diez S, Aisen P, et al. (2002) Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infect Immun 70: 5124–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nosanchuk JD, van Duin D, Mandal P, Aisen P, Legendre AM, et al. (2004) Blastomyces dermatitidis produces melanin in vitro and during infection. FEMS Microbiol Lett 239: 187–193. [DOI] [PubMed] [Google Scholar]
- 42. Tsai HF, Fujii I, Watanabe A, Wheeler MH, Chang YC, et al. (2001) Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J Biol Chem 276: 29292–29298. [DOI] [PubMed] [Google Scholar]
- 43. de Hoog GS, Vicente V, Caligiorne RB, Kantarcioglu S, Tintelnot K, et al. (2003) Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J Clin Microbiol 41: 4767–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Hoog GS, Attili-Angelis D, Vicente VA, Gerrits van den Ende AH, Queiroz-Telles F (2004) Molecular ecology and pathogenic potential of Fonsecaea species. Med Mycol 42: 405–416. [DOI] [PubMed] [Google Scholar]
- 45. Kumar SV, Phale PS, Durani S, Wangikar PP (2003) Combined sequence and structure analysis of the fungal laccase family. Biotechnol Bioeng 83: 386–394. [DOI] [PubMed] [Google Scholar]
- 46. Kawasaki M, Aoki M, Ishizaki H, Miyaji M, Nishimura K, et al. (1999) Molecular epidemiology of Fonsecaea pedrosoi using mitochondrial DNA analysis. Med Mycol 37: 435–440. [PubMed] [Google Scholar]
- 47. Tanabe H, Kawasaki M, Mochizuki T, Ishizaki H (2004) Species identification and strain typing of Fonsecaea pedrosoi using ribosomal RNA gene internal transcribed spacer regions. Nihon Ishinkin Gakkai Zasshi 45: 105–112. [DOI] [PubMed] [Google Scholar]