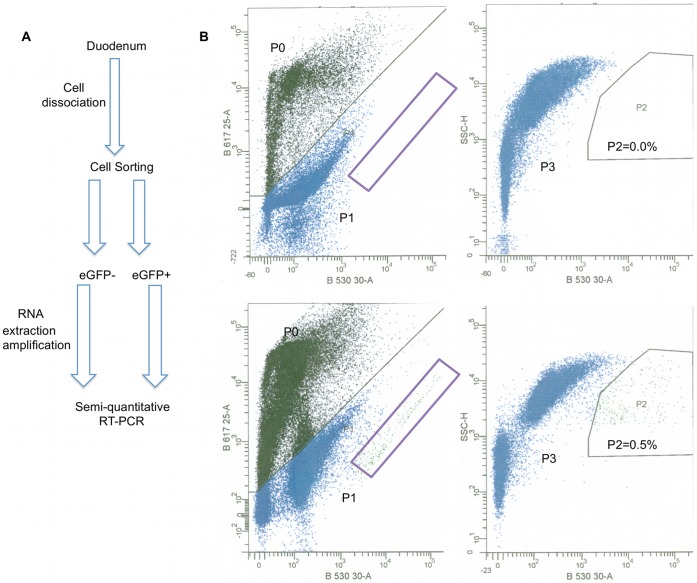
Figure 2. FACS-sorting of eGFP+ and eGFP− cells.
A: Schematic overview of the methods employed to isolate I-cells and perform transcript analysis. B: Dissociated duodenal cells from eGFP-CCK mice were sorted using a BD FACS Aria cell sorter (lower panels). Dissociated duodenal cells from a WT/CD-1 mouse were used as a control to set gating parameters for analysis and sorting (upper panels). Cells were first stained with PI and analyzed by eGFP (X-axis) and PI (Y-axis) fluorescence intensity (left panels). PI- positive cells (dead cells, P0) were excluded from further analysis. Live cells (Blue, P1) from CCK-eGFP mice displayed a population of highly fluorescent cells (purple rectangle) that were not present in cell from WT mice. Live cells were also analyzed based on eGFP fluorescence intensity (X-axis) and side scatter (Y-axis). Cells that expressed eGFP were gated as eGFP+ cells, representing 0.5% of total cell population (P2, right lower panel). This population was not present in cells from WT mice (P2, right upper panel). All other cells (P3) were gated as eGFP−. eGFP+ cells and approximately equal number of eGFP− cells were sorted.