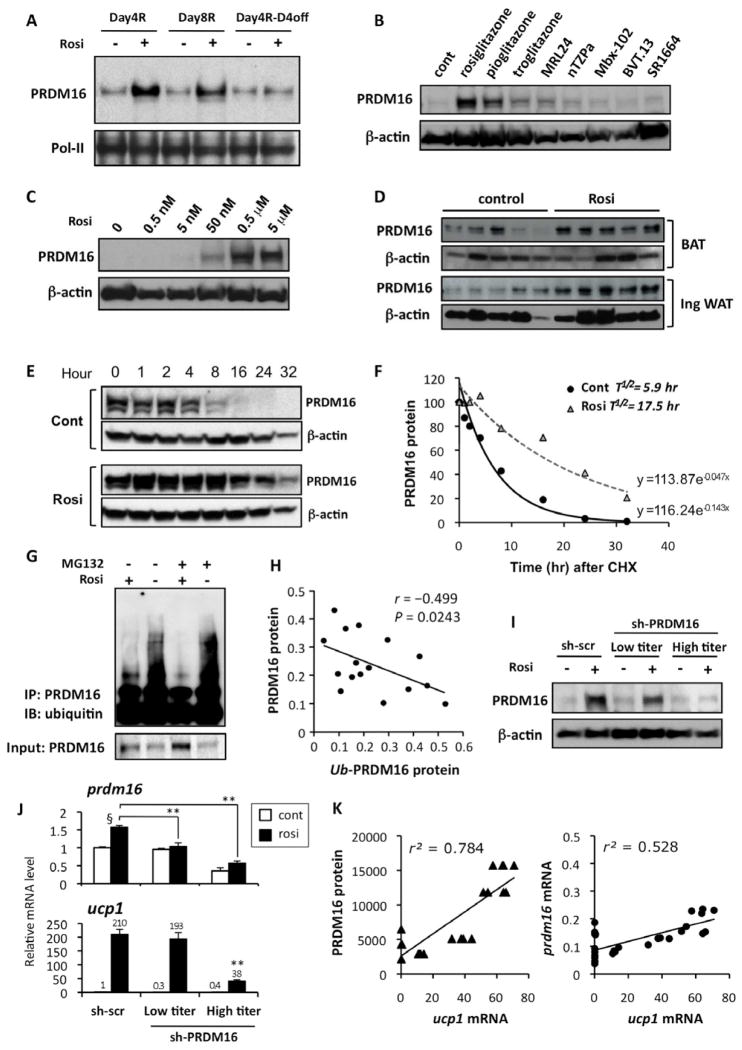
Fig. 4. Browning of the white adipocytes is tightly linked to the rosiglitazone-induced PRDM16 protein stabilization.
(A) Primary inguinal cells from SVF were differentiated in the presence or absence of rosiglitazone at 1μM for 4 days (Day4R), for 8 days (Day8R), and for 4 days with rosiglitazone and additional 4 days without rosiglitazone (Day4R-D4off). PRDM16 protein levels were analyzed by Western blotting. Pol-II was blotted as a loading control. (B) Primary inguinal WAT-derived adipocytes were treated with full or partial agonists for PPARγ. PRDM16 protein levels were analyzed by Western blotting. β-actin was blotted as a loading control. (C) PRDM16 protein levels were analyzed by Western blotting in primary adipocytes treated with rosiglitazone at different doses. (D) PRDM16 protein levels were analyzed by Western blotting in BAT and inguinal WAT isolated from wild-type mice. Mice were injected IP with saline or rosiglitazone at 10mg/kg for 10 days. (E) PRDM16 protein levels in a cycloheximide chase experiment were analyzed by Western blotting. Top panel: control cells, Bottom panel: rosiglitazone treated cells (F) Regression analysis of PRDM16 protein stability in (E). (G) F442A cells expressing flag-tagged PRDM16 were differentiated in the presence or absence of rosiglitazone. Cells were treated with a proteasome inhibitor MG132 (10 μM) for 16 hours prior to harvesting the cells. PRDM16 was immunoprecipitated using flag antibody, and ubiquitin was detected by Western blotting. Total PRDM16 protein is shown in the bottom panel. (H) Correlation analysis between PRDM16 protein levels and ubiquitinated PRDM16 levels. (I) PRDM16 protein levels were analyzed by Western blotting in primary inguinal adipocytes infected with control scramble shRNA or shRNA targeting PRDM16 at low- or high-tires adenovirus. (J) mRNA levels for prdm16 and ucp1 were measured by qRT-PCR in (I). **P<0.01 relative to sh-scr control cells treated with rosiglitazone. § P<0.01 relative to cells without rosiglitazone treatment. (K) Correlation analyses between ucp1 mRNA levels and PRDM16 protein levels (left panel), and between ucp1 mRNA levels and prdm16 mRNA levels (right panel). Data are expressed as means ± SEM.