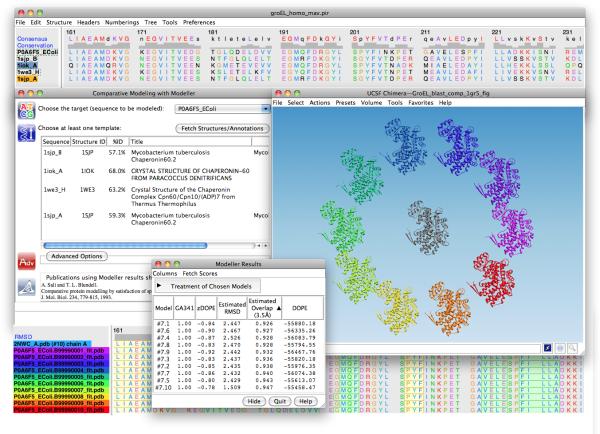
Figure 1.
Chimera MODELLER interface and homology models of a monomer of E. coli GroEL. A multiple sequence alignment of the target with hits from a Chimera BLAST query is displayed in Multalign Viewer (top window). In the Comparative Modeling with Modeller dialog (left), the P0A6F5_EColi (GroEL) sequence is selected as the target and sequences 1sjp_A, 1sjp_B, 1iok_A and 1we3_H are designated as templates. The resulting modeled structures are shown in a circle around the experimentally determined structure of E. coli GroEL (PDB: 2NWC) in the UCSF Chimera 3D graphics window. The sequence alignment between the models and 2NWC is also included (bottom window), as are the modeling scores (Modeller Results window).