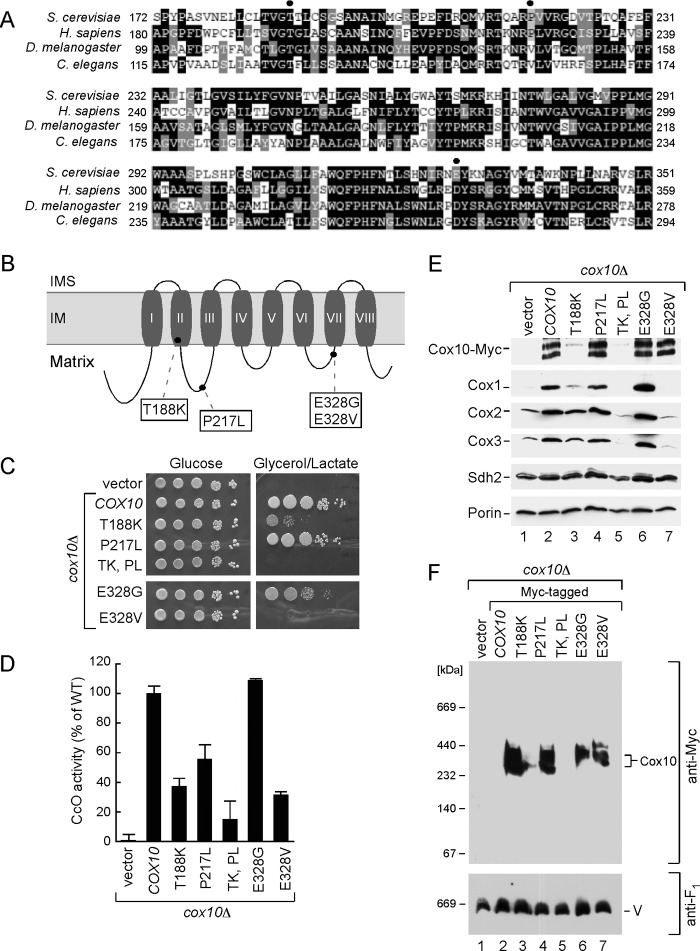
FIGURE 6.
Analysis of Cox10 human pathogenic mutations in the yeast model. A, multiple sequence alignment of the Cox10 conserved region from different species. Sequences were aligned using the MultAlin and BoxShade programs. Identical amino acid residues are shown in black; conserved residues are in dark gray, and the similar ones are in light gray. The circles indicate amino acid residues mutated in Leigh syndrome patients. B, schematic view of yeast Cox10. Predicted transmembrane domains (I–VIII) are indicated. Marked are the substitutions corresponding to pathogenic mutations in human protein. C, respiratory growth of wild-type (WT) and cox10Δ cells of BY4743 background expressing WT Cox10 or its T188K, P217W, T188K/P217W, E328G, and E328V mutant forms. Cells were handled and tested as in Fig. 2C, except that glycerol/lactate plates were incubated up to 6 days at 30 °C. D, mitochondria derived from the cells described in C were used to assess CcO activity. Enzymatic activities are shown as a percentage of WT-specific activity. The data represent an average of three independent measurements; the error bars indicate S.D. E, steady-state levels of Cox10-13Myc and its mutant forms as well as core CcO subunits in the respective mitochondria (10 μg) were analyzed by immunoblot with indicated antibodies. Subunit Sdh2 of succinate dehydrogenase and outer membrane protein porin were detected with respective antibodies and served as controls. F, BN-PAGE analysis of the aforementioned mitochondria. Samples were handled and analyzed as in Fig. 2.