Background: Some membrane proteins, such as Kir2.3, are internalized by a clathrin-dependent mechanism even though they lack canonical endocytic signals.
Results: A new endocytic signal in Kir2.3, consisting of two pairs of hydrophobic residues, interacts within a hydrophobic cleft in AP-2 ασ2 subunits.
Conclusion: The repertoire of signals recognized by AP-2 is wider than previously anticipated.
Significance: This study provides new insights into the mechanism of clathrin-mediated endocytosis.
Keywords: Adaptor Proteins, Endocytosis, Ion Channels, Membrane Proteins, Membrane Trafficking, Potassium Channels, Trafficking, AP-2, Kir Channels, Kir2.3
Abstract
Select plasma membrane proteins can be marked as cargo for inclusion into clathrin-coated pits by common internalization signals (e.g. YXXΦ, dileucine motifs, NPXY) that serve as universal recognition sites for the AP-2 adaptor complex or other clathrin-associated sorting proteins. However, some surface proteins, such as the Kir2.3 potassium channel, lack canonical signals but are still targeted for clathrin-dependent endocytosis. Here, we explore the mechanism. We found an unusual endocytic signal in Kir2.3 that is based on two consecutive pairs of hydrophobic residues. Characterized by the sequence ΦΦXΦΦ (a tandem di-hydrophobic (TDH) motif, where Φ is a hydrophobic amino acid), the signal shows no resemblance to other endocytic motifs, yet it directly interacts with AP-2 to target the Kir2.3 potassium channel into the endocytic pathway. We found that the tandem di-hydrophobic motif directly binds to the ασ2 subunits of AP-2, interacting within a large hydrophobic cleft that encompasses part of the docking site for di-Leu signals, but includes additional structures. These observations expand the repertoire of clathrin-dependent internalization signals and the ways in which AP-2 can coordinate endocytosis of cargo proteins.
Introduction
By controlling the density of specific subsets of receptors, ion channels, and transporters at the plasma membrane, clathrin-mediated endocytosis influences numerous physiologic and developmental processes. The AP-2 clathrin adaptor complex has a central role in clathrin-mediated endocytosis. By simultaneously interacting with specific surface proteins and clathrin, AP-2 couples cargo selection to clathrin-coated pit formation. To date, two different cargo recognition mechanisms have been described. Many clathrin-mediated endocytosis-targeted proteins contain a common endocytic signal (i.e. either YXXΦ or di-Leu ((DE)XXXL(LIM)) (1–3)) that directly interacts with distinct binding pockets in AP-2. In other cases, AP-2 acts in an indirect capacity, binding to other clathrin-associated sorting proteins, which in turn directly interact with different endocytic signals on cargo proteins (i.e. NPXY or ubiquitin) (4) through well defined protein-protein binding mechanisms. However, some clathrin-mediated endocytosis-internalized proteins do not contain a canonical internalization signal (5–10), raising a question regarding the mechanism by which AP-2 can facilitate their endocytosis.
Different AP-2 cargo recognition mechanisms are illustrated by the inwardly rectifying K+ (Kir) channels, a group of structurally related membrane proteins that play important roles in neuronal excitability, hormone secretion, heart rate, and salt balance (11–16). For example, AP-2 directly interacts with Kir6.2 in a YXXΦ signal-dependent manner to target this ATP-sensitive potassium channel for internalization (17). By contrast, a clathrin-associated sorting protein, ARH, binds simultaneously to AP-2 and to an NPXY-type motif in Kir1.1 to recruit these channels into clathrin-coated pits (18, 19). In other cases, the endocytic mechanism remains unclear. For example, an unusual di-isoleucine-containing sequence in Kir2.3 is required for the channel to be internalized via a clathrin- and AP-2-dependent pathway (10). Although the internalization sequence shares some similarities with the di-Leu signal, the sequence requirements of the Kir2.3 signal appear to be different compared with the canonical motif, and it has been a mystery as to how the endocytic signal is recognized by AP-2.
Separate binding sites for the canonical signals have been identified in AP-2. The adaptor complex is composed of a small σ2 subunit (∼17 kDa), an intermediate μ subunit (∼47 kDa), and two large subunits (α and β, ∼100 kDa). YXXΦ motifs are well appreciated to bind to the μ subunit (20, 21). The binding site for dileucine signals proved to be much more elusive, but with the advent of approaches to study recombinant AP-2 in vitro, the docking structure has been recently mapped to a site within the σ2 and α subunits (2, 3, 22–24). In this study, we used a similar approach to fully characterize the endocytic motif in Kir2.3 and map its binding site in AP-2.
EXPERIMENTAL PROCEDURES
DNA Constructs, Plasmids, and Antibodies
Bicistronic DNA constructs in pFastBac Dual, encoding the ασ2 adaptin hemicomplexes, were kindly provided by Dr. Stuart Kornfeld (Washington University School of Medicine, St. Louis, MO) (23). GST-tagged mouse Kir2.3 (amino acids 340–445) and Nef (full-length) fusion proteins were cloned into pGEX-4T1 (GE Healthcare). CD4 and CD4-Kir2.3 (amino acids 340–445) chimeras and full-length Kir2.3 containing an external FLAG epitope tag subcloned in pcDNA3.1(+) (Invitrogen) have been described previously (10). The antibodies used and their sources were as follows: mouse anti-HA (16B12, Covance), mouse anti-Myc and agarose-conjugated mouse anti-Myc (9B11, Cell Signaling Technology), mouse anti-adaptin-α (BD Transduction Laboratories), and rabbit anti-adaptin-α (Sigma). Rabbit anti-Kir2.3 antibody was kindly provided by Dr. D. S. Bredt (Department of Physiology, University of California, San Francisco, CA) (25). The eluting peptides used were HA and FLAG (Sigma).
Protein Expression, Purification, and Pulldown Assays
GST, GST-Nef, and GST-Kir2.3 fusion proteins were produced in bacteria, affinity-purified, dialyzed in PBS, and quantified. Infectious baculovirus particles encoding adaptin hemicomplexes were produced as described by Doray et al. (23) using the Bac-to-Bac baculovirus expression system (Invitrogen). Sf9 cells expressing the recombinant AP subunits were lysed in buffer containing 25 mm HEPES-KOH (pH 7.2), 125 mm potassium acetate, 2.5 mm magnesium acetate, 2 mm dithiothreitol, and 0.4% Triton X-100 supplemented with EDTA-free protease inhibitor mixture (Roche Applied Science). For GST affinity chromatography studies, 25 μg of GST fusion protein was bound to glutathione-Sepharose beads, incubated with 50 μl of Sf9 lysate (0.8 μg/μl) for 1 h at 4 °C, and then washed three times for 15 min at 4 °C with lysis buffer containing 2% Triton X-100. Specifically bound hemicomplexes were resolved by SDS-PAGE and visualized by Western blotting using anti-epitope antibodies and a SNAP i.d.® detection system (Millipore).
Immunoprecipitation of Adaptin Hemicomplexes
Sf9 cell lysates (supplemented with 2% BSA) were incubated with agarose-conjugated anti-Myc antibodies, washed with PBS containing 2% BSA and 1% Triton X-100, washed with PBS containing 1% Triton X-100, eluted with SDS protein sample buffer, and subjected to Western blotting with an anti-HA and anti-Myc antibodies.
Co-immunoprecipitation of AP-2 and Kir2.3
Cell lysates of HEK293 cells in buffer A (20 mm HEPES (pH 7.6), 25 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Triton X-100, and protease inhibitors) were rotated overnight with anti-α monoclonal antibodies and Protein G Plus-Protein A beads. Beads were washed four times with PBS and then eluted for 30 min at room temperature with SDS sample buffer. Immunoprecipitated AP-2 and Kir2.3 were visualized by SDS-PAGE and Western blotting using appropriate polyclonal antibodies.
Biotinylation Internalization Assay
Cells were washed with ice-cold Ringer's solution (5 mm HEPES, 144 mm NaCl, 5 mm KCl, 1.2 mm NaH2PO4, 5.5 mm glucose, 1 mm MgCl2, and 1 mm CaCl2 (pH 7.4)), and the surface proteins were biotinylated with EZ-Link sulfo-NHS-SS-biotin (0.3 mg/ml; Pierce) in Ringer's solution at 4 °C for 15 min. The remaining biotin was quenched for 20 min at 4 °C with 50 mm Tris (pH 7.5) in Ringer's solution. Cells were then placed at 37 °C for the indicated amount of time (0–10 min) to allow internalization. MesNa (100 mm in 100 mm NaCl, 1 mm EDTA, 0.2% BSA, and 50 mm Tris (pH 8.8)) was added three times for 20 min at 4 °C to cleave biotin linked to the cell surface. Cells were then washed with Ringer's solution and lysed in buffer A. 50 μg of total protein was incubated overnight at 4 °C with NeutrAvidin beads (Pierce), washed with 1% Triton X-100 in PBS, and eluted from the beads with SDS sample buffer, and biotinylated Kir2.3 was quantified by dot blotting (Bio-Rad) using anti-Kir2.3 antibody.
Antibody-feeding Internalization Assay
COS-7 cells were studied 48 h after transfection with the CD4-Kir2.3 chimeras (FuGENE 6, Roche Applied Science). CD4-Kir2.3 proteins at the plasma membrane of live cells were labeled with an Alexa Fluor 488-conjugated anti-CD4 antibody that recognizes an external epitope on CD4 for 1 h at 4 °C in low glucose DMEM. Unbound antibody was washed with DMEM, and cells were rapidly raised to 37 °C to permit internalization for 5 min. After returning the cells to 4 °C, the remaining surface-bound antibody was stripped with DMEM and 100 mm glycine (pH 3.0). Cells were washed with PBS, fixed in 4% paraformaldehyde, mounted, and visualized using a Zeiss confocal laser scanning microscope (63× oil immersion lens, numerical aperture of 1.40). Internalized CD4 proteins, visualized as intracellular puncta, were measured (NIH ImageJ 1.40g) and quantified as pixel area relative to the cell surface area.
Structural Models and Statistics
Structural models based on available Protein Data Bank files were visualized using PyMOL (DeLano Scientific LLC). Data (means ± S.E.) were subjected to one-way analysis of variance and the Bonferroni post hoc test (GraphPad Prism).
RESULTS
Kir2.3 Interacts with ασ2 Subunits of AP-2
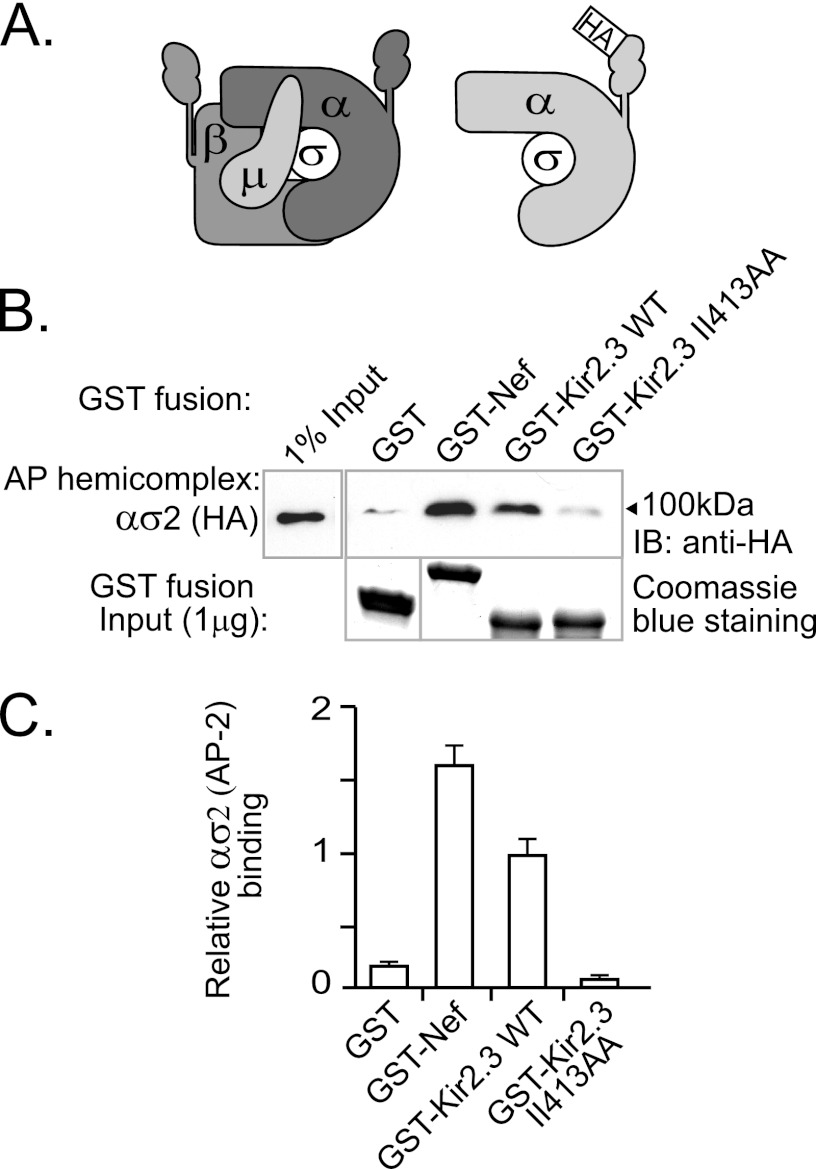
To determine how the endocytic signal in Kir2.3 interacts with clathrin adaptors, in vitro binding studies with recombinant AP-2 subunits were performed. Because expression of the AP-2 complex as heterodimers (ασ2 and β2μ2) generates appropriately folded hemicomplexes that have exposed binding sites for di-Leu or YXXΦ motifs (23), we felt that this approach might be optimal to map the recognition site for the unique endocytic motif in Kir2.3. AP-2 ασ2 subunits were coexpressed in Sf9 cells and were made detectable with an HA epitope tag in the larger α subunit (Fig. 1A). We hypothesized that the Kir2.3 di-isoleucine may behave similarly to dileucine motifs and consequently bind to the ασ2 heterodimer. To test for interaction with Kir2.3, a GST-Kir2.3 fusion protein containing the di-isoleucine-based motif was used as an affinity ligand in pulldown experiments with ασ2 adaptin hemicomplexes (Fig. 1, B and C). A GST fusion protein of the full-length HIV protein Nef, which contains a canonical di-Leu signal, served as a binding control (22, 26). As detected by Western blotting with anti-HA (α) antibodies, GST-Kir2.3 interacted with the ασ2 hemicomplexes of AP-2. The mutation II413AA, decreased binding to near-background levels, indicating that the interaction of Kir2.3 with ασ2 is dependent on residues required for endocytosis.
FIGURE 1.
Binding of recombinant epitope-tagged adaptin hemicomplexes to the AP-2 ασ2 hemicomplex. A, schematic of the AP-2 complex (left) together with an HA epitope-tagged AP-2 hemicomplex (right). B, Western blots of ασ2 adaptin hemicomplexes (upper panel) produced in Sf9 cells and pulled down with immobilized GST-Kir2.3 or GST-Nef fusion proteins (lower panel). Hemicomplexes strongly bound to Nef and Kir2.3. Minimal binding was detected with either GST alone or GST fused to Kir2.3 mutant II413AA. IB, immunoblot. C, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.).
The AP-2 Interaction Signal in Kir2.3 Accepts Leucine in Place of Isoleucine
Sequence analysis of di-Leu-based endocytic signals in other transmembrane proteins reveals that isoleucine commonly replaces the second leucine (L+1), but only in rare occasions does it substitute for the L(L0) residue. Until Kir2.3, it had never been found to replace both leucines simultaneously (1). Given our observation that the di-isoleucine motif in Kir2.3 interacts with the same AP-2 subunits as di-Leu motifs, we wondered if the Kir2.3 signal is an atypical variant of the canonical di-Leu signal or a completely new and unrelated endocytic motif that interacts with AP-2 ασ2 through a different mechanism. To begin to test this, we used a mutagenesis approach to determine whether the sequence requirements for Kir2.3 interaction with AP-2 ασ2 are similar to or different from those for the canonical di-Leu signal in Nef.
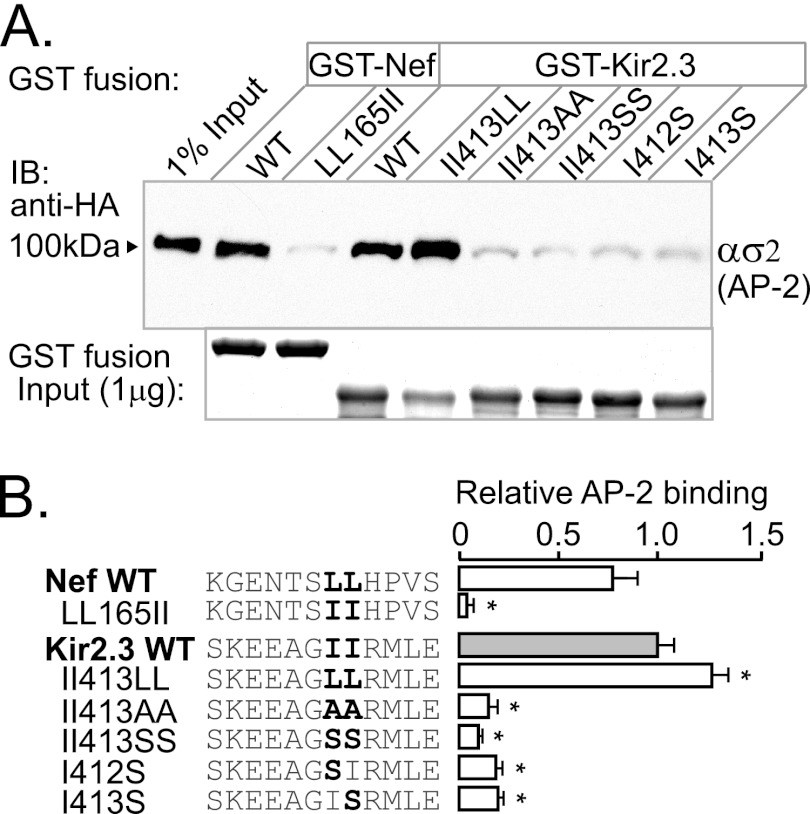
As measured in in vitro pulldown experiments, replacement of the isoleucines in the Kir2.3 endocytic signal with leucines (II413LL) did not attenuate the interaction with AP-2 ασ2 (Fig. 2). By contrast, AP-2 ασ2 interaction with Nef was completely inhibited by the reciprocal mutation of dileucine to di-isoleucine (LL165II). Thus, the requirements of specific hydrophobic residues in the Kir2.3 signal are more flexible compared with the canonical signal. Nevertheless, it does display a key similarity. Like di-Leu signals, Kir2.3 does not support interaction with AP-2 ασ2 when the comparable hydrophobic residues (di-isoleucine) are replaced with smaller hydrophobic (Ala) or polar (Ser) residues. The general di-hydrophobic requirements of the interaction signal suggest that Kir2.3 may bind to a hydrophobic pocket in the AP-2 hemicomplex, like the di-Leu signals.
FIGURE 2.
Endocytic motif in Kir2.3 accepts Leu in place of Ile. To investigate the specific requirement for isoleucine in the Kir2.3 motif, the residues were replaced with leucine, alanine, or serine, and the mutant forms were tested for ασ2 interaction in GST pulldown assays. A, representative Western blots (upper panel) and Coomassie Blue staining of GST fusion proteins (lower panel). IB, immunoblot. B, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05). The II413LL mutation modestly increased binding to ασ2, whereas LL165II in Nef completely prevented binding. By contrast, the II413SS mutation decreased binding, similar to II413AA. Individual substitution of Kir2.3 Ile412 or Ile413 with serine blocked interaction, revealing that both Ile0 and Ile+1 are essential for binding.
Common Binding Site in AP-2
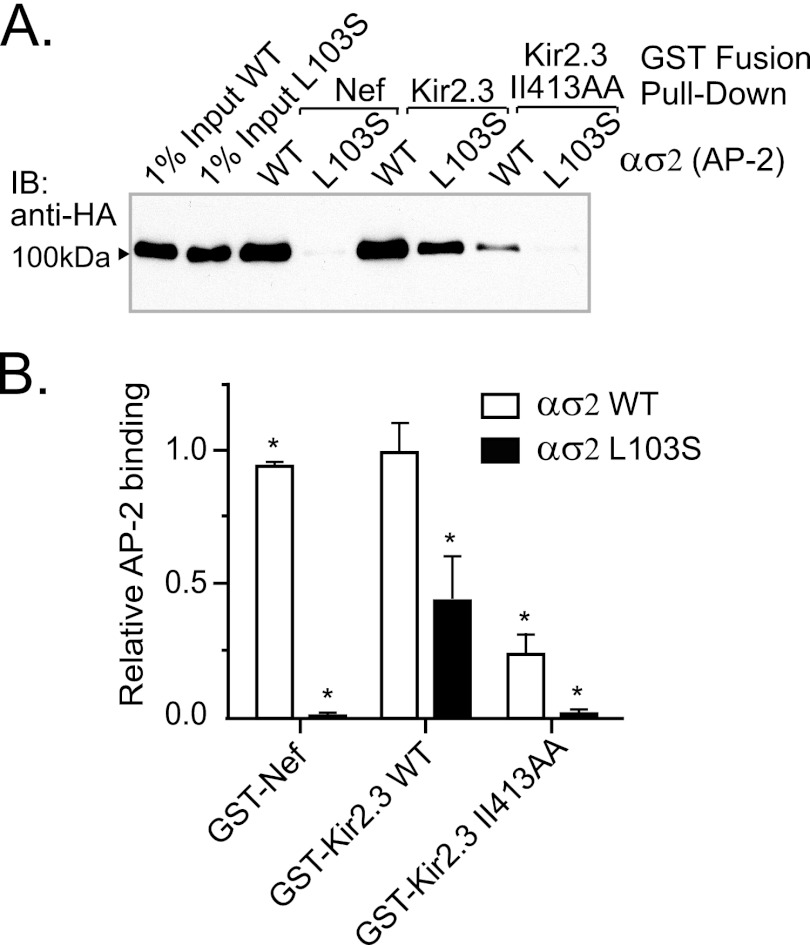
To determine whether Kir2.3 interacts with AP-2 at the same site as the canonical signal, a structure-directed mutagenesis approach was employed, guided by the recent solution of the AP-2 adaptor core atomic structure in complex with a di-Leu signal (from CD4) (24). For initial analysis, we selected a specific residue in the di-Leu-binding pocket of the AP-2 σ2 subunit, σ2Leu103, because it lies in close proximity to the first leucine of the interacting endocytic di-Leu signal, and it has been shown to govern interaction with the CD4 peptide. As measured in pulldown studies, mutation of this residue to serine completely blocked interaction of the ασ2 subunit with Nef (Fig. 3), corroborating the absolute necessity of σ2Leu103 for interaction with the di-Leu signal. By comparison, the σ2L103S mutation caused a large but incomplete reduction in binding to WT Kir2.3 (66% (S.E. 16%), p < 0.01), suggesting that the Kir2.3-binding site in AP-2 shares some overlap with the canonical di-Leu-binding site. Taken together with the observation that residual interaction of the Kir2.3 II413AA mutant with AP-2 ασ2 was completely eliminated by the σ2L103S mutation, we reasoned that additional structures in Kir2.3 and AP-2 may also contribute to the interaction.
FIGURE 3.
Nef and Kir2.3 share a common binding site in AP-2. A, mutation of a key residue in the di-Leu-binding pocket of AP-2 (σ2L103S) prevented binding to Nef and significantly decreased binding to Kir2.3. Shown are representative Western blots. IB, immunoblot. B, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05).
The AP-2 Recognition Signal in Kir2.3 Includes Hydrophobic Amino Acids downstream of Di-Ile413
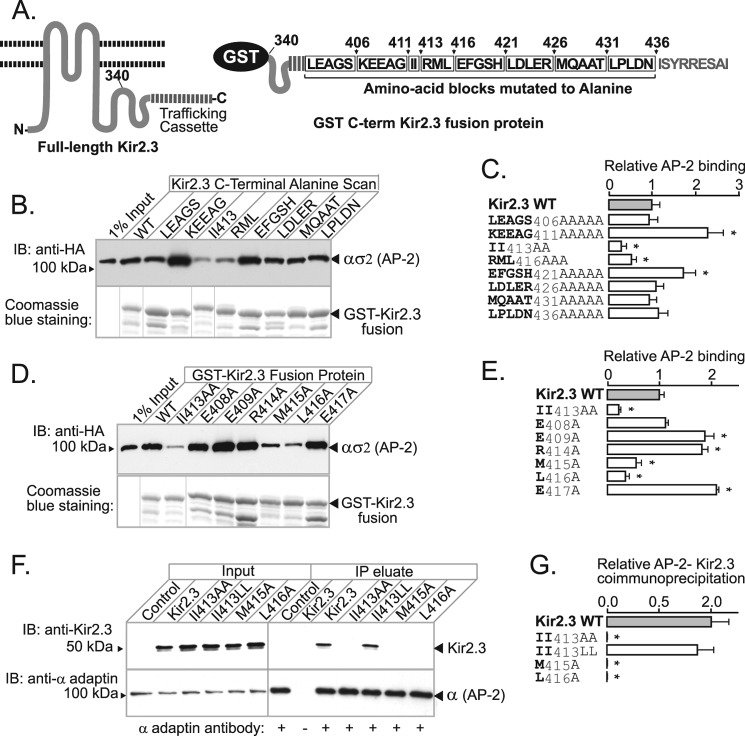
To identify additional amino acids in Kir2.3 that contribute to AP-2 interaction, an alanine scanning mutagenesis approach was combined with the in vitro binding assay (Fig. 4A). The initial round of mutagenesis was done in blocks, replacing strings of consecutive residues that surround di-Ile413 with alanine (Fig. 4, B and C). This pinpointed tracks of residues that either are required for binding (RML416) or impede interaction (KEEAG411 and EFGSH421). Point mutations within these tracks revealed the residues that are absolutely required for AP-2 binding (Met415 and Leu416, in addition to di-Ile413) and those than have a negative contribution (Glu409, Arg414, and Glu417) (Fig. 4, D and E). As measured by co-immunoprecipitation in HEK293 cells (Fig. 4, F and G), the requirements for Kir2.3 interaction with AP-2 were found to be identical in vivo; alanine replacement of di-Ile413 or ML416 in Kir2.3 abolished its ability to interact with AP-2 in cells. Thus, the AP-2-binding sequence spans a 5-amino acid linear stretch, characterized by a tandem di-hydrophobic (TDH)2 motif (IIXML416).
FIGURE 4.
Hydrophobic residues in Kir2.3 located downstream of di-Ile413 contribute to AP-2 binding. A, schematic representation of the C terminus of Kir2.3 expressed as a GST fusion protein (left), highlighting blocks of residues subjected to alanine scanning mutagenesis (right). B, representative binding study. Mutants are identified by residues replaced with alanine. AP-2 ασ2 bound to the WT and each of the mutant fusion proteins (lower panel) was accessed in Western blotting with anti-HA antibody (upper panel). IB, immunoblot. C, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05). D, representative binding study of point mutants. E, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05). F, identical hydrophobic residues are required for the interaction and co-immunoprecipitation of Kir2.3 expressed in HEK293 cells with endogenous AP-2. IP, immunoprecipitate. G, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05).
AP-2 Residues αArg21 and σ2Arg15 Are Not Required for TDH Motif Binding
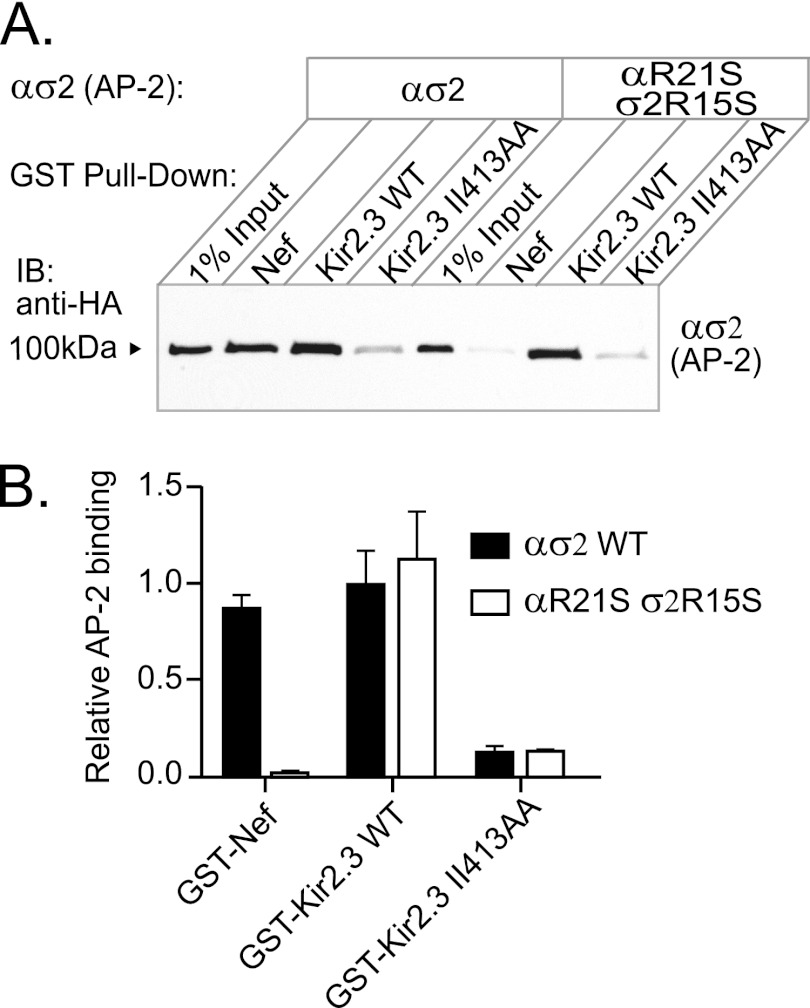
The interaction of canonical dileucine-binding signals depends on a negatively charged residue at position L-4 ((DE)XXXL(LIM)), which has been shown to interact with a positively charged patch constituted by both αArg21 and σ2Arg15 (24). Because the TDH motif does not contain a similar negatively charged residue, we hypothesized that it would interact with AP-2 independently of the positively charged patch. To test this, we mutated the critical patch residues (αArg21/σ2Arg15) to serine and tested the mutant heterodimers in pulldown assays for binding with Nef and Kir2.3 (Fig. 5). As expected, mutant hemicomplexes were unable to bind Nef, but binding to Kir2.3 was unaffected, indicating that this area of ασ2 plays no role in Kir2.3 interaction.
FIGURE 5.
αArg21 and σ2Arg15 are not involved in Kir2.3 binding. The positively charged residues that interact with Asp or Glu in a canonical di-Leu motif are contributed by αArg21 and σ2Arg15 in AP-2. A, to determine whether these amino acids contribute to binding of the TDH motif in Kir2.3, hemicomplexes containing the mutations αR21S and σ2R15S were used in pulldown assays with GST-Kir2.3. Mutant hemicomplexes were competent for Kir2.3 binding but were unable to interact with Nef. IB, immunoblot. B, densitometric quantification and summary of the data from three separate experiments (mean ± S.E., p < 0.05).
Val104 and Phe105 Contribute to the TDH Motif-binding Structure
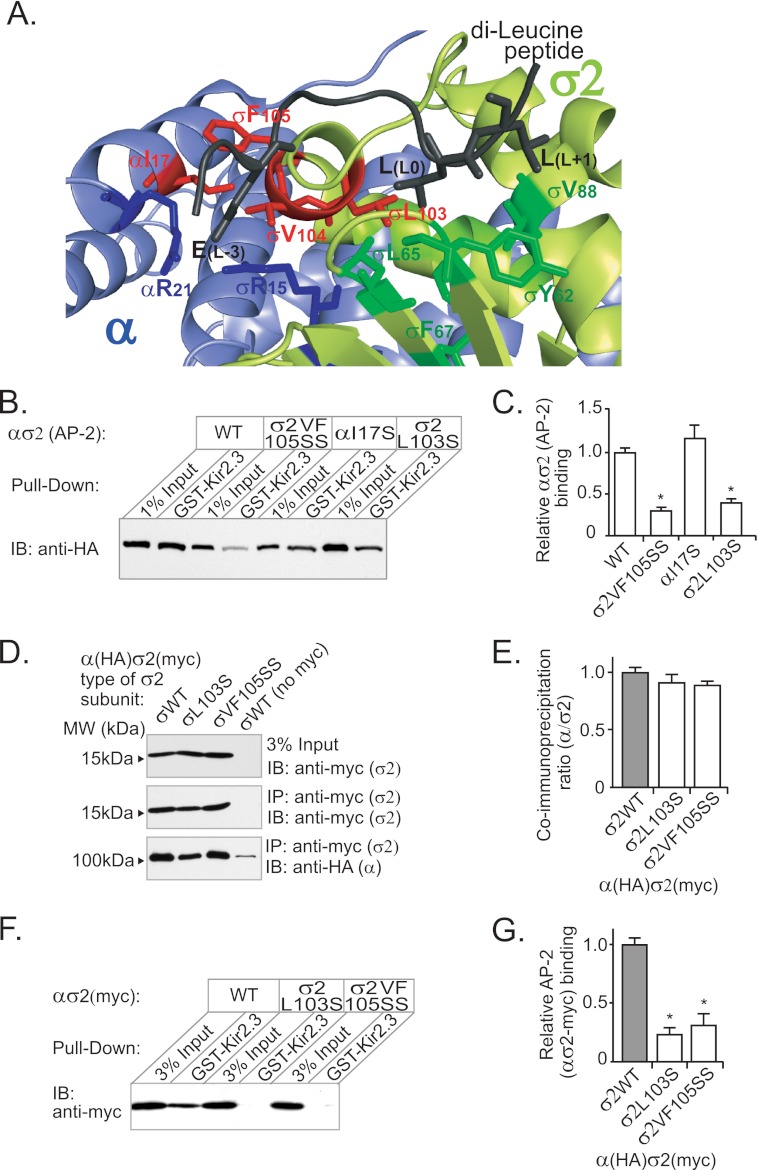
The hydrophobic binding pocket for canonical di-Leu signals, as presently defined (24), composed of Leu103, Leu65, Phe67, Tyr62, and Val88 in AP-2 σ2 (Fig. 6A), is too small to simultaneously accommodate both pairs of hydrophobic amino acids in the Kir2.3 internalization motif (Fig. 6A and supplemental Fig. S2). Accordingly, candidate interacting structures were identified in the atomic structure of AP-2 as hydrophobic residues within a 22-Å radius (the predicted length of the most extended conformation of the IIRML peptide) of σ2Leu103 and tested as the additional determinants of Kir2.3 binding. Serine or alanine replacement mutations of hydrophobic residues at the outer edge of the potential interaction interface (αTyr250, αPhe251, and αVal300) had no effect on Kir2.3 binding (supplemental Fig. S3). By contrast, several residues in a proximal cluster of hydrophobic amino acids, composed of αIle17, σ2Phe105, and σ2Val104, were found to be required for Kir2.3 interaction. Indeed, serine replacement mutations at σ2Phe105 and σ2Val104 (VF105SS), but not at αIle17, dramatically reduced AP-2 ασ2 interaction with Kir2.3 (Fig. 6, B and C).
FIGURE 6.
Val104 and Phe105 in the AP-2 σ2 subunit contribute to the Kir2.3-binding site. A, model of AP-2 in complex with a di-Leu peptide from CD4 (Gln-to-Glu mutant) based on Protein Data Bank code 2JKT (24). The hydrophobic side chains that participate in the di-Leu motif-binding site (Tyr62, Leu65, Phe67, and Val88 in dark green and Leu103 in red) are contributed by the σ2 subunit (pale green). Additional hydrophobic amino acids (σ2Val104, σ2Phe105, and αIle17 in red) are clustered to one side of the di-Leu-binding site. B, to determine whether these amino acids contribute to an extended hydrophobic pocket capable of accommodating the larger Kir2.3 internalization motif, hemicomplexes containing the mutations σ2VF105SS and αI17S were used in pulldown assays with GST-Kir2.3. σ2VF105SS decreased binding as much as σL103S, whereas αI17S had no effect on binding. IB, immunoblot. C, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05). D, Myc-tagged wild-type and mutant σ2 subunits were expressed as hemicomplexes with HA-tagged α subunits and subjected to immunoprecipitation (IP) with anti-Myc monoclonal antibody. The HA-tagged α subunit co-immunoprecipitated with equal efficiency with wild-type and mutant σ2 subunits (detected by Western blotting using anti-HA and anti-Myc antibodies), showing that mutation of σ2 did not affect the overall assembly of the hemicomplexes. No co-immunoprecipitation was observed with control untagged σ2 hemicomplexes. E, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.). F, pulldown assay with GST-Kir2.3 using hemicomplexes containing WT and mutant Myc-tagged σ2 subunits also displayed decrease binding to σ2L103S and σ2VF105SS. G, densitometric quantification and summary of the data from three separate experiments (mean ± S.E.; *, p < 0.05).
In principle, the σ2L103S or σ2VF105SS mutations might affect Kir2.3 interaction indirectly by altering the assembly or stability of hemicomplexes. To rule out this possibility, we generated hemicomplexes that contained a C-terminal Myc-tagged σ2 subunit, in addition to the HA-tagged α subunit, and performed co-immunoprecipitation studies to test for complex assembly. As shown, σ2 mutant subunits (either L103S or VF105SS) co-assembled with the α subunit equally well as with the WT σ2 subunit (Fig. 6, D and E). Furthermore, pulldown experiments with GST-Kir2.3 and double-tagged hemicomplexes, using Myc-σ2 as the input control, produced similar results as those with hemicomplexes containing only one tag (HA tag in α) and using HA-α as the input control (Fig. 6, F and G). Thus, the σ2L103S and σ2VF105SS mutations did not significantly affect subunit oligomerization, providing strong evidence that the mutation does not impair global structure. Taken together, these observations indicate the AP-2 hydrophobic binding pocket is larger than previously imagined, allowing interaction of the TDH internalization motif in Kir2.3.
TDH Motif Is Required for Kir2.3 Endocytosis
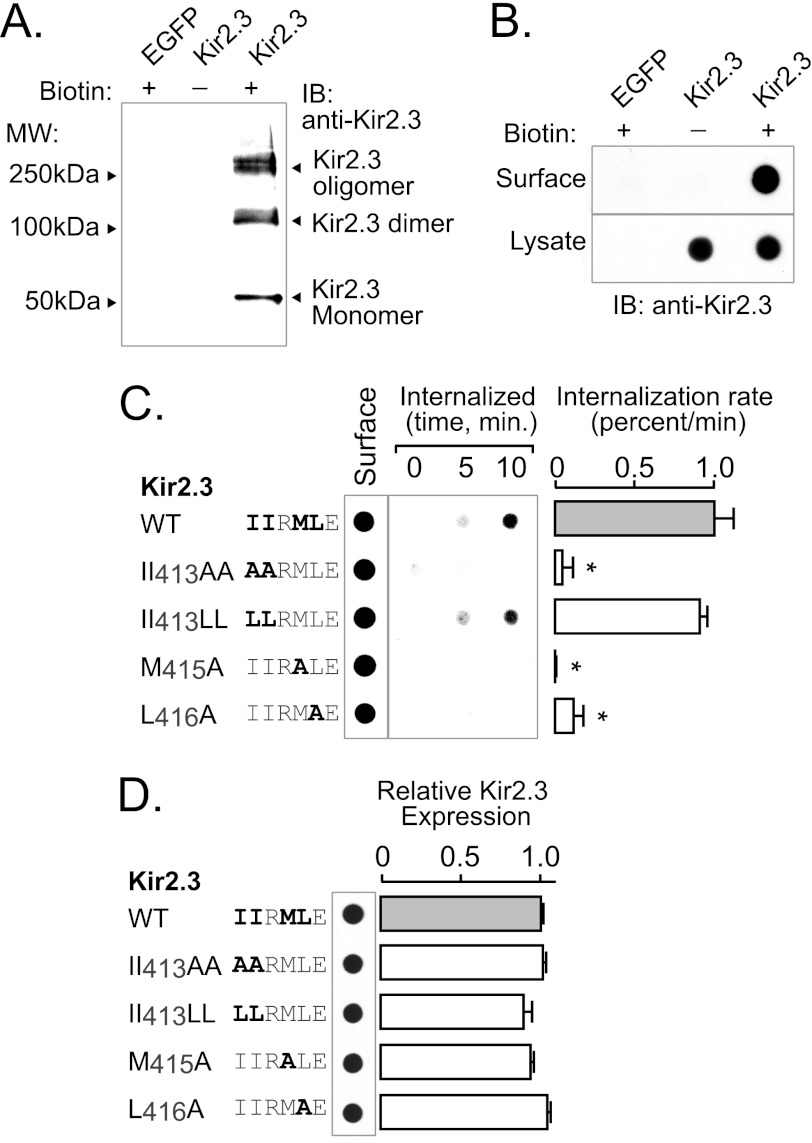
To determine whether residues in the extended AP-2 interaction motif participate in internalization, we measured the endocytic response to disruption of the binding residues (Fig. 7). For these studies, WT and mutant Kir2.3 channels were transfected into HEK293 cells, and the internalization rate was evaluated using a biotinylation assay as described under “Experimental Procedures.” Because biotinylated Kir2.3 subunits often migrated as monomeric and multimeric species on SDS-polyacrylamide gels (Fig. 7A), we used a dot blot assay to collectively detect all biotinylated forms of Kir2.3 with a single densitometric measurement for more reliable quantification. As shown in Fig. 7B, biotinylated Kir2.3 was specifically detected using this method. Using this approach to measure internalization of biotinylated Kir2.3 from the cell surface, we found that the hydrophobic residues in Kir2.3 that are necessary for AP-2 binding are also required for endocytosis (Fig. 7C). Mutation of each hydrophobic amino acid in the Kir2.3 TDH motif resulted in a significant reduction of the Kir2.3 internalization rate (Fig. 7C), without effects on the total abundance of the channel (Fig. 7D).
FIGURE 7.
TDH motif (IIXML416) is required for internalization of Kir2.3. Rates of endocytosis were determined in HEK293 cells expressing Kir2.3 (WT or mutant) using a surface biotin labeling/internalization chase assay. A, surface-biotinylated Kir2.3 was specifically detected in Western blots with anti-Kir2.3 antibodies. Shown is avidin-bound material from HEK293 cells transfected with Kir2.3 or enhanced GFP (EGFP) and surface-labeled with a cell-impermeant biotin derivative (+) or vehicle (−). Biotinylated Kir2.3 migrated on SDS-polyacrylamide gels as three species: a 50-kDa (monomeric) band, a 100-kDa (dimeric) band, and a higher molecular mass oligomeric form. Identical molecular mass forms were specifically detected in Western blots of whole cell lysates of Kir2.3-transfected HEK293 cells (data not shown). B, dot-immunoblot analysis of Kir2.3 with anti-Kir2.3 antibodies specifically detected the biotinylated channel at the cell surface. Avidin-purified material was compared with the whole lysate of cells transfected with Kir2.3 or enhanced GFP and surface-labeled with biotin (+) or vehicle (−). In contrast to Kir2.3-transfected cells, nothing was detected in cells transfected with Kir2.3 and labeled with vehicle or in enhanced GFP-transfected cells labeled with biotin, verifying specificity. C, a representative biotin internalization experiment is shown (left panel), next to the summary data (right panel). In these studies, surface channels were labeled with the cell-impermeant biotin derivative in the cold and then incubated at 37 °C for 0, 5, or 10 min to capture the initial rate of endocytosis. Biotin remaining at the cell surface was cleaved with MesNa, and internalized (biotinylated) Kir2.3 was recovered and detected along with the total surface pool in dot immunoblots. The internalization rate of mutant II413AA, M415A, and L416A channels was greatly reduced compared with WT and mutant II413LL Kir2.3 channels (mean ± S.E.; *, p < 0.05). IB, immunoblot. D, by contrast, mutation of the hydrophobic residues involved in AP-2 binding and endocytosis did not affect total Kir2.3 protein abundance.
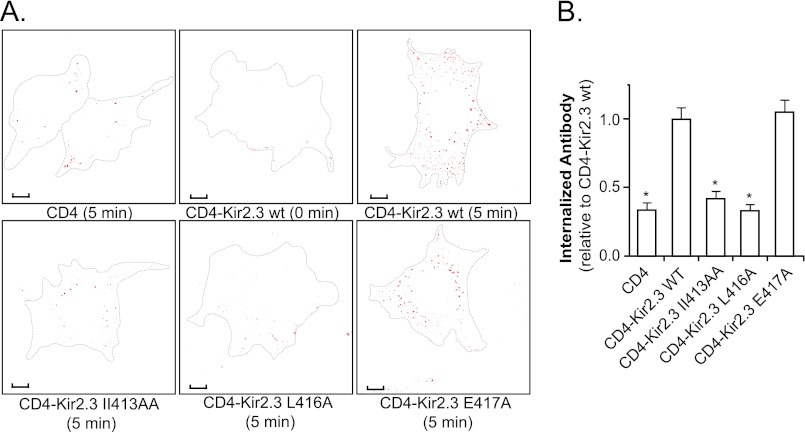
To determine whether the TDH motif acts as an autonomous internalization signal, a CD4-Kir2.3 chimera was constructed using the extracellular and transmembrane domains of CD4 and a C-terminal fragment of Kir2.3 that includes the TDH motif. Because biotin is very difficult to remove from the large extracellular domain of CD4, endocytosis was assessed using an antibody-feeding assay as described under “Experimental Procedures.” In these studies, chimeric proteins on the cell surface were labeled with anti-CD4 antibody in COS-7 cells and allowed to internalize over a 5-min chase. After all anti-CD4 antibody remaining at the surface was stripped away, the internalized chimera was visualized in endosomes by confocal microscopy and quantified (Fig. 8, A and B). Using this approach, we found that the residues in the TDH motif were absolutely required for endosomal accumulation. By contrast, mutation of a nearby residue (E417A), which did not inhibit AP-2 interaction, had no effect on endosomal targeting. Taken together, these observations indicate that the TDH motif operates as an autonomous internalization signal.
FIGURE 8.
TDH motif acts as an autonomous internalization signal. A, shown are representative COS-7 cells transfected with CD4 or the CD4-Kir2.3 chimeras (WT or mutant) at 0 and 5 min post-endocytic chase and after anti-CD4 antibodies were stripped from the cell surface. Scale bars = 10 μm. Accumulation of the surface-labeled chimera in endosomes was reduced to background levels by alanine replacement mutations in the di-hydrophobic motif. B, summary of the internalization data of CD4 and CD4-Kir2.3 chimeras, documenting the number of internalized antibody-labeled particles relative to the WT CD4-Kir2.3 chimera (mean ± S.E., n = 10; *, p < 0.05).
DISCUSSION
Here, we have uncovered an AP-2 adaptor signal recognition mechanism. Extending our analysis of Kir2.3 channel endocytosis (10) to a comprehensive screen of residues required for AP-2 interaction, an endocytic signal was defined. Composed of two consecutive pairs of hydrophobic residues (ΦΦxΦΦ, where Φ may be Ile, Leu, or Met), the TDH internalization signal diverges from other endocytic signals. Importantly, it also differs in the way it associates with AP-2. A hydrophobic binding cleft on the σ2 subunit of the tetrameric AP-2 adaptor complex was identified as the TDH motif-docking site. Although parts of the binding pocket are shared with canonical di-Leu signals (24), the TDH motif interacts with AP-2 differently from the well recognized motif.
The docking structure in σ2 for the TDH motif not only encompasses the hydrophobic binding site for the canonical di-Leu signal (involving Leu103, Tyr62, Leu65, and Val88) but also includes an adjacent hydrophobic patch. As resolved by structure-guided mutagenesis, the TDH interaction site extends a least ∼14 Å beyond Leu103 to include σ2Val104 and σ2Phe105 (Fig. 6A and supplemental Fig. S2). Viewed in the atomic structure of AP-2 (24), these intervening hydrophobic residues are spaced at distances that are predicted to favor strong interactions with hydrophobic peptides (27), creating a binding pocket that is optimal for interaction with the TDH motif. Although we were able to establish that the hydrophobic binding cleft in AP-2 is large enough to accommodate a TDH motif, it is important to appreciate the limitation of our approach. Because of the long-range nature of hydrophobic interactions, it is not possible to precisely map the binding site in AP-2 for each specific hydrophobic residue of the TDH motif by mutagenesis. Such a precise description of the interaction of AP-2 with a TDH signal at the atomic level will likely require more crystallographic studies. Nonetheless, all residues involved in formation of the TDH hydrophobic binding pocket, as defined by our approach, are maintained from Drosophila to man (supplemental Fig. S1A), consistent with an evolutionarily conserved pocket for signal recognition. Moreover, similar endocytosis motifs are present in other channels. For example, the dileucine cluster (HLLDLLDE) clathrin-dependent internalization signal in the ClC-3 chloride channel (28, 29) is predicted to interact with AP-2 in a similar manner as Kir2.3. Thus, it appears that the hydrophobic cleft in the ασ subunits of AP-2 is equipped to bind multiple types of hydrophobic endocytic signals, facilitating internalization of a variety of cargo proteins.
Another key difference in the way the TDH signal is recognized by AP-2 compared with the canonical di-Leu signal is highlighted by the contrasting requirements of electrostatic interactions. Optimal recognition of the di-Leu signal requires pairing between its negatively charged amino acid at position L-4 ((DE)XXXL(LI)) and basic residues (αArg21 and σ2Arg15) in the AP-2 ασ2-binding site, although there are some exceptions (1, 30, 31). By contrast, our mutational analysis of the Kir2.3 internalization signal revealed that charged residues at the equivalent position (Glu409) or flanking positions (Arg414 or Glu417) are not required for AP-2 interaction or endocytosis. In fact, the electrostatic binding pocket in AP-2 (αArg21/σ2Arg15) could also be mutated without compromising AP-2 binding to the TDH motif in Kir2.3. Based on these observations, it is reasonable to propose that the TDH motif docks into a single large hydrophobic pocket rather than into a two-slotted electrostatic and hydrophobic socket, like the (DE)XXXL(LI) signal. As a consequence, interaction of a TDH signal with AP-2 should be governed largely by favorable hydrophobic binding energies.
Although the binding sites for di-Leu and TDH signals are different, they possess enough overlap to be controlled through a common regulatory mechanism. Recent crystallographic studies reveal that AP-2 undergoes a large conformational change upon association with phosphatidylinositol 4,5-bisphosphate-containing membranes, causing the (ED)XXXL(LI)-binding site to become exposed (32). The proximity of the TDH motif-binding site suggests that it is also blocked by parts of the β2 subunit in the closed conformation and would also require phosphatidylinositol 4,5-bisphosphate binding to become accessible for interaction. Also like the di-Leu-docking site, the TDH motif-docking structure is predicted to be in the same plane as the phosphatidylinositol 4,5-bisphosphate-binding site. This would allow AP-2 to simultaneously engage transmembrane cargo (containing TDH signals) and phosphatidylinositol 4,5-bisphosphate-containing membranes.
This study, revealing the TDH motif, corroborates and extends our previous identification of the di-isoleucine part of the internalization signal in Kir2.3 (10). Indeed, each hydrophobic amino acid that was identified in the TDH signal as essential for direct AP-2 binding was also found to be necessary for rapid Kir2.3 internalization. It should be pointed out that the previous assessment of channel accumulation in endosomes, using an antibody-feeding internalization assay with a reporter construct (10), suggested that endocytosis may not supported by replacement of the di-isoleucine residues (di-Ile413) with dileucine. In this study, we measured endocytosis more directly with the entire channel, rather than a reporter, over a faster time frame and found that, unlike the II413AA mutation, the II413LL mutation did not affect channel endocytosis. The finding is consistent with the requirements for AP-2 binding and provides support for our conclusion that the endocytic signal in Kir2.3 is based on two consecutive pairs of hydrophobic residues.
In summary, we have identified an AP-2 adaptor signal recognition mechanism, expanding the repertoire of ways that cell surface proteins can be recognized as cargo for clathrin-dependent endocytosis.
Acknowledgments
We thank Stuart Kornfeld and Balraj Dorian for providing the DNA constructs containing the wild-type adaptin hemicomplexes in pFastBac Dual and for technical advice in the production of the hemicomplexes.
This work was supported, in whole or in part, by National Institutes of Health Grants DK63049 and DK54231. This work was also supported the National Kidney Foundation of Maryland.

This article contains supplemental Figs. S1–S3.
- TDH
- tandem di-hydrophobic.
REFERENCES
- 1. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 2. Owen D. J., Collins B. M., Evans P. R. (2004) Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 20, 153–191 [DOI] [PubMed] [Google Scholar]
- 3. Traub L. M. (2009) Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10, 583–596 [DOI] [PubMed] [Google Scholar]
- 4. Welling P. A., Weisz O. A. (2010) Sorting it out in endosomes: an emerging concept in renal epithelial cell transport regulation. Physiology 25, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leser G. P., Ector K. J., Ng D. T., Shaughnessy M. A., Lamb R. A. (1999) The signal for clathrin-mediated endocytosis of the paramyxovirus SV5 HN protein resides at the transmembrane domain-ectodomain boundary region. Virology 262, 79–92 [DOI] [PubMed] [Google Scholar]
- 6. Blagoveshchenskaya A. D., Hewitt E. W., Cutler D. F. (1999) Dileucine signals mediate targeting of tyrosinase and synaptotagmin to synaptic-like microvesicles within PC12 cells. Mol. Biol. Cell 10, 3979–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao M., Gold L., Dorward H., Liang L. F., Hoodbhoy T., Boja E., Fales H. M., Dean J. (2003) Mutation of a conserved hydrophobic patch prevents incorporation of ZP3 into the zona pellucida surrounding mouse eggs. Mol. Cell. Biol. 23, 8982–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanemoto M., Abe T., Ito S. (2005) PDZ-binding and di-hydrophobic motifs regulate distribution of Kir4.1 channels in renal cells. J. Am. Soc. Nephrol. 16, 2608–2614 [DOI] [PubMed] [Google Scholar]
- 9. Yokode M., Pathak R. K., Hammer R. E., Brown M. S., Goldstein J. L., Anderson R. G. (1992) Cytoplasmic sequence required for basolateral targeting of LDL receptor in livers of transgenic mice. J. Cell Biol. 117, 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mason A. K., Jacobs B. E., Welling P. A. (2008) AP-2-dependent internalization of potassium channel Kir2.3 is driven by a novel di-hydrophobic signal. J. Biol. Chem. 283, 5973–5984 [DOI] [PubMed] [Google Scholar]
- 11. Bichet D., Haass F. A., Jan L. Y. (2003) Merging functional studies with structures of inward rectifier K+ channels. Nat. Rev. Neurosci. 4, 957–967 [DOI] [PubMed] [Google Scholar]
- 12. Nichols C. G., Lopatin A. N. (1997) Inward rectifier potassium channels. Annu. Rev. Physiol. 59, 171–191 [DOI] [PubMed] [Google Scholar]
- 13. Karschin C., Dissmann E., Stühmer W., Karschin A. (1996) IRK(1–3) and GIRK(1–4) inwardly rectifying K+ channel mRNAs are differentially expressed in the adult rat brain. J. Neurosci. 16, 3559–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melnyk P., Zhang L., Shrier A., Nattel S. (2002) Differential distribution of Kir2.1 and Kir2.3 subunits in canine atrium and ventricle. Am. J. Physiol. Heart Circ. Physiol. 283, H1123–H1133 [DOI] [PubMed] [Google Scholar]
- 15. Millar I. D., Taylor H. C., Cooper G. J., Kibble J. D., Robson L. (2006) A Kir2.3-like K+ conductance in mouse cortical collecting duct principal cells. J. Membr. Biol. 211, 173–184 [DOI] [PubMed] [Google Scholar]
- 16. Welling P. A. (1997) Primary structure and functional expression of a cortical collecting duct Kir channel. Am. J. Physiol. 273, F825–F836 [DOI] [PubMed] [Google Scholar]
- 17. Mankouri J., Taneja T. K., Smith A. J., Ponnambalam S., Sivaprasadarao A. (2006) Kir6.2 mutations causing neonatal diabetes prevent endocytosis of ATP-sensitive potassium channels. EMBO J. 25, 4142–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang L., Garuti R., Kim B. Y., Wade J. B., Welling P. A. (2009) The ARH adaptor protein regulates endocytosis of the ROMK potassium secretory channel in mouse kidney. J. Clin. Invest. 119, 3278–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng W. Z., Babich V., Ortega B., Quigley R., White S. J., Welling P. A., Huang C. L. (2002) Evidence for endocytosis of ROMK potassium channel via clathrin-coated vesicles. Am. J. Physiol. Renal Physiol. 283, F630–F639 [DOI] [PubMed] [Google Scholar]
- 20. Ohno H., Stewart J., Fournier M. C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., Bonifacino J. S. (1995) Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 [DOI] [PubMed] [Google Scholar]
- 21. Owen D. J., Evans P. R. (1998) A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaudhuri R., Lindwasser O. W., Smith W. J., Hurley J. H., Bonifacino J. S. (2007) Down-regulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP-2 clathrin adaptor. J. Virol. 81, 3877–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doray B., Lee I., Knisely J., Bu G., Kornfeld S. (2007) The γ/σ1 and α/σ2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol. Biol. Cell 18, 1887–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly B. T., McCoy A. J., Späte K., Miller S. E., Evans P. R., Höning S., Owen D. J. (2008) A structural explanation for the binding of endocytic dileucine motifs by the AP-2 complex. Nature 456, 976–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen N. A., Brenman J. E., Snyder S. H., Bredt D. S. (1996) Binding of the inward rectifier K+ channel Kir2.3 to PSD-95 is regulated by protein kinase A phosphorylation. Neuron 17, 759–767 [DOI] [PubMed] [Google Scholar]
- 26. Doray B., Knisely J. M., Wartman L., Bu G., Kornfeld S. (2008) Identification of acidic dileucine signals in LRP9 that interact with both GGAs and AP-1/AP-2. Traffic 9, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Israelachvili J., Pashley R. (1982) The hydrophobic interaction is long range, decaying exponentially with distance. Nature 300, 341–342 [DOI] [PubMed] [Google Scholar]
- 28. Zhao Z., Li X., Hao J., Winston J. H., Weinman S. A. (2007) The ClC-3 chloride transport protein traffics through the plasma membrane via interaction of an N-terminal dileucine cluster with clathrin. J. Biol. Chem. 282, 29022–29031 [DOI] [PubMed] [Google Scholar]
- 29. Stauber T., Jentsch T. J. (2010) Sorting motifs of the endosomal/lysosomal CLC chloride transporters. J. Biol. Chem. 285, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pitcher C., Höning S., Fingerhut A., Bowers K., Marsh M. (1999) Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol. Biol. Cell 10, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sandoval I. V., Martinez-Arca S., Valdueza J., Palacios S., Holman G. D. (2000) Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 275, 39874–39885 [DOI] [PubMed] [Google Scholar]
- 32. Jackson L. P., Kelly B. T., McCoy A. J., Gaffry T., James L. C., Collins B. M., Höning S., Evans P. R., Owen D. J. (2010) A large-scale conformational change couples membrane recruitment to cargo binding in the AP-2 clathrin adaptor complex. Cell 141, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]