FIG. 4.
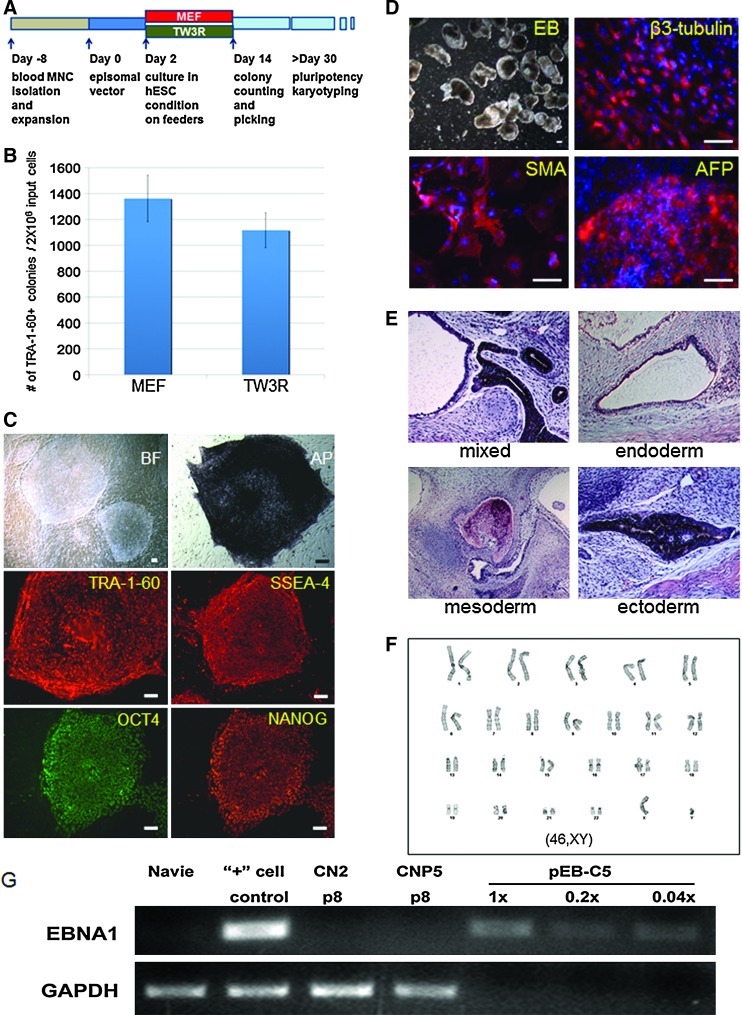
Efficient derivation of integration-free human iPSCs on TW3R human feeders. (A) Schematics of reprogramming blood cells with a single episomal vector. Un-fractionated cord blood MNCs were expanded for 8 days in erythroblast culture condition before being transfected by a single EBNA1/OriP based episomal vector expressing 5 reprogramming factors. After 2 days of recovery, the transfected cells were plated onto MEF or TW3R plates for reprogramming. iPS-like TRA-1-60 positive colonies were counted 14 days after episomal vector transfection. (B) Both MEF and TW3R support efficient generation of TRA-1-60+ colonies. After colony picking and expansion for 6–8 passages, 2 of the candidate iPS lines (CN2 and CNP5) generated on TW3R were fully characterized. (C) CN2 expresses typical pluripotency-related cell surface markers and transcription factors. (D) Immunofluorescence staining of beta-3-tubulin (ectoderm marker), SMA (mesoderm marker), and AFP (endodermal marker) in EBs and (E) Representative tissue types from endoderm (respiratory epithelium), mesoderm (bone), and ectoderm (pigmented epithelium) in teratoma demonstrated the pluripotency of the CN2 cell line. (F) The CN2 iPS line also showed a normal male karyotype (46,XY) after 7 passages. (G) Vector DNA was not detected in the CN2 or CNP5 cell line by PCR analysis specifically for EBNA1 sequence. The single-copy cellular sequence GAPDH was used as a quality control for isolated genomic DNA. Total DNA isolated from the cells before (naïve) and after nucleofection (day 2, “+” cell control) was used as negative and positive controls, respectively. The episomal plasmid diluted in amounts equivalent to 1, 0.2, or 0.04 copies per genome of cellular DNA was also used as controls for the detection of the EBNA1 DNA sequence. MNC, mononuclear cell; PCR, polymerase chain reaction; GAPDH, glyceraldehyde 3-phophate dehydrogenase. Color images available online at www.liebertonline.com/scd