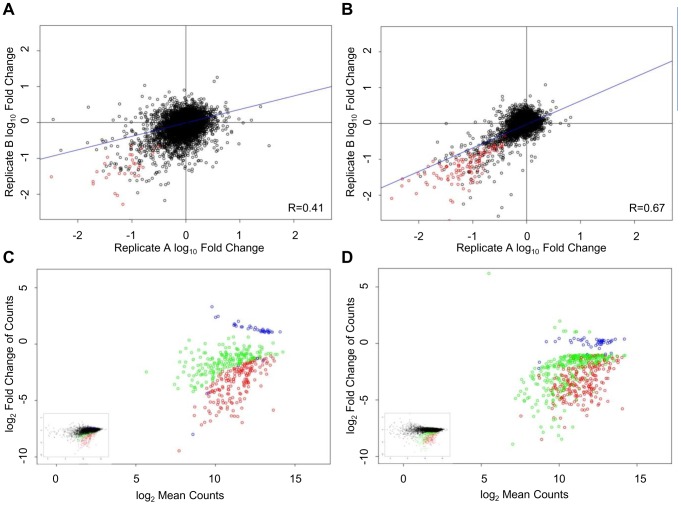
Figure 4. Effect of fold representation of shRNA at transduction on HEK293T viability screen reproducibility using NGS analysis.
Viability screens performed in Figure 3 analyzed by NGS. Scatter plot of log10(T1/T0) of the biological replicates of the S100 (A) and S500 (B) screens are shown with Pearson correlation values indicated in the corner of each plot. Primary hits (shRNA that passed fold change criteria of (T1/T0) greater than two and FDR rate of ≤0.05 in both screening (biological) replicates) are depicted in red. Signal (log2Mean Counts) for S100 (C) and S500 (D) screens are plotted as a function of log ratio (log 2(T1/T0)). Primary hits are color coded with hits identified in both S100 and S500 screens (red), hits identified in the S100 screen only (blue) and hits identified in the S500 screen only (green). The complete data set is presented in the small insert.