Table 1. Sensitivity analysis percentages using various methods (data from Mikkelsen et al. 2007 [8]).
| unit-mean | quantile | MACS | ChIPDiff | rank | RSEG | two-stage unit-mean | ChIPnorm | |
| thresholds |
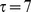
|
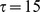
|
p-val 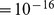
|
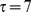
|

|
cdf 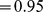
|
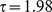
|

|
| NP differential (four-fold) expressed: 925 genes | ||||||||
| sensitivity (ES K27-enriched) | 14.49 | 15.24 | 31.24 | 14.64 | 17.62 | 18.16 | 15.14 | 15.14 |
| error (NP K27-enriched) | 0 | 0 | 0 | 0.27 | 6.05 | 0.11 | 0 | 0 |
| ES differential (four-fold) expressed: 1104 genes | ||||||||
| sensitivity (NP K27-enriched) | 0 | 1.27 | 0.27 | 0 | 12.59 | 1.9 | 6.88 | 7.16 |
| error (ES K27-enriched) | 1.27 | 1.99 | 5.07 | 0.91 | 5.43 | 3.08 | 1.18 | 1.99 |
Experiments: unit-mean; quantile; MACS peak finder; ChIPDiff; Rank normalization; two-stage unit-mean; ChIPnorm. The parameters of all the methods (except MACS) were adjusted so that all of them give almost the same percentage (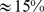 ) of experiment “sensitivity (ES K27-enriched)”.
) of experiment “sensitivity (ES K27-enriched)”.
